주메뉴
- About IBS 연구원소개
-
Research Centers
연구단소개
- Research Outcomes
- Mathematics
- Physics
- Center for Underground Physics
- Center for Theoretical Physics of the Universe (Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe (Cosmology, Gravity and Astroparticle Physics Group)
- Dark Matter Axion Group
- Center for Artificial Low Dimensional Electronic Systems
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Exotic Nuclear Studies
- Center for Van der Waals Quantum Solids
- Center for Relativistic Laser Science
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research (Neuro Technology Group)
- Center for Neuroscience Imaging Research (Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Genome Engineering
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Institutes
- Korea Virus Research Institute
- News Center 뉴스 센터
- Career 인재초빙
- Living in Korea IBS School-UST
- IBS School 윤리경영


주메뉴
- About IBS
-
Research Centers
- Research Outcomes
- Mathematics
- Physics
- Center for Underground Physics
- Center for Theoretical Physics of the Universe (Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe (Cosmology, Gravity and Astroparticle Physics Group)
- Dark Matter Axion Group
- Center for Artificial Low Dimensional Electronic Systems
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Exotic Nuclear Studies
- Center for Van der Waals Quantum Solids
- Center for Relativistic Laser Science
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research (Neuro Technology Group)
- Center for Neuroscience Imaging Research (Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Genome Engineering
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Institutes
- Korea Virus Research Institute
- News Center
- Career
- Living in Korea
- IBS School
News Center
|
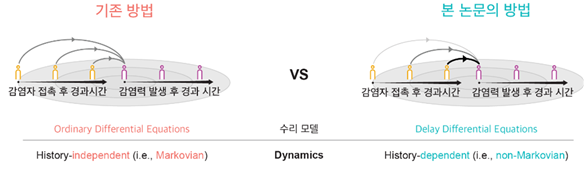
What Was Wrong with Traditional Epidemic Models? — “Models That Forgot the Past”One of the key indicators describing how fast an infectious disease spreads is the effective reproduction number (R-value), which plays a decisive role in shaping public health policies. However, most widely used epidemic-spread models so far have relied on an unrealistic assumption that all infected individuals transmit the disease with a constant probability. This assumption corresponds to a Markov process, in which the probability of future infection depends only on the current state, without considering when in the past the infection occurred. In reality, however, the infectiousness of a disease changes over time — for example, an individual’s infectiousness may increase or decrease depending on how much time has passed since infection. This means real-world transmission follows a non-Markovian process more closely. The mismatch can distort estimates of the reproduction number, especially during the early, rapidly changing phase of an outbreak. Understanding the Reproduction NumberThe reproduction number (R) indicates how many additional people one infected person infects on average at a given time, making it essentially the “driving force” of an epidemic. If R exceeds 1, the outbreak grows; if it is below 1, it fades. Because R can show how interventions such as testing, isolation, or vaccination affect transmission early on, it serves as a key policy indicator. However, R is not a simple ratio — it is sensitive to when transmissions occur (the time distribution between infection and secondary transmission). If this distribution is not modeled realistically, R may spike unrealistically high during the initial phase of an epidemic. Therefore, when interpreting R, one must consider realistic generation intervals, as well as policy changes, reporting delays, testing volume, and under-reporting. In short, to make R a reliable compass for public-health decisions, models must capture not only how many transmissions occur but also when they occur. Estimation Based on a Non-Markovian Infection ModelTransmission evolves over time: right after infection, contagiousness is low; it peaks at a certain time and then gradually declines. The non-Markovian approach incorporates this time dependence directly into the model. Instead of assuming a simple exponential distribution, it uses more realistic distributions for infection and incubation periods (for example, a gamma distribution) to represent when infectiousness rises and how long it lasts. By combining this with Bayesian inference, the model can estimate key epidemiological variables — such as the reproduction number and infectious period — with high accuracy even from case counts alone. This makes it extremely useful for real-time epidemic-control planning.
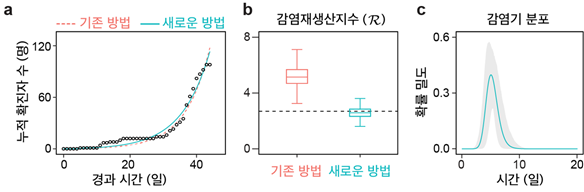
Re-evaluating COVID-19 DataApplying this method to early-stage COVID-19 data from Korea showed that traditional models (which assume a fixed infectious period) overestimated R at around 4, whereas the new non-Markovian approach yielded R ≈ 2.7 — consistent with actual contact-tracing data. This demonstrates that how a model handles the timing of transmissions can change the interpretation of the same dataset. Moreover, this approach can estimate the infectious-period distribution itself, allowing policymakers to quantitatively assess the impact of interventions such as testing and isolation.
Toward More Refined Policy DecisionsBy providing more stable and realistic estimates of R, the new method reflects real changes in the transmission timeline before and after interventions. As a result, it can reduce excessive fluctuations in epidemic-peak forecasts and resource-demand projections. The team’s open-source software package also enables customized simulations that include factors such as vaccination, viral variants, asymptomatic infections, and fatality rates — offering strong potential to improve both the precision and speed of policy design for future emerging infectious diseases. |
| before |
|---|
- Content Manager
- Communications Team : Kwon Ye Seul 042-878-8237
- Last Update 2023-11-28 14:20