주메뉴
- About IBS 연구원소개
-
Research Centers
연구단소개
- Research Outcomes
- Mathematics
- Physics
- Center for Underground Physics
- Center for Theoretical Physics of the Universe (Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe (Cosmology, Gravity and Astroparticle Physics Group)
- Dark Matter Axion Group
- Center for Artificial Low Dimensional Electronic Systems
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Exotic Nuclear Studies
- Center for Van der Waals Quantum Solids
- Center for Relativistic Laser Science
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research (Neuro Technology Group)
- Center for Neuroscience Imaging Research (Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Genome Engineering
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Center for Quantum Conversion Research
- Institutes
- Korea Virus Research Institute
- News Center 뉴스 센터
- Career 인재초빙
- Living in Korea IBS School-UST
- IBS School 윤리경영


주메뉴
- About IBS
-
Research Centers
- Research Outcomes
- Mathematics
- Physics
- Center for Underground Physics
- Center for Theoretical Physics of the Universe (Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe (Cosmology, Gravity and Astroparticle Physics Group)
- Dark Matter Axion Group
- Center for Artificial Low Dimensional Electronic Systems
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Exotic Nuclear Studies
- Center for Van der Waals Quantum Solids
- Center for Relativistic Laser Science
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research (Neuro Technology Group)
- Center for Neuroscience Imaging Research (Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Genome Engineering
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Center for Quantum Conversion Research
- Institutes
- Korea Virus Research Institute
- News Center
- Career
- Living in Korea
- IBS School
News Center
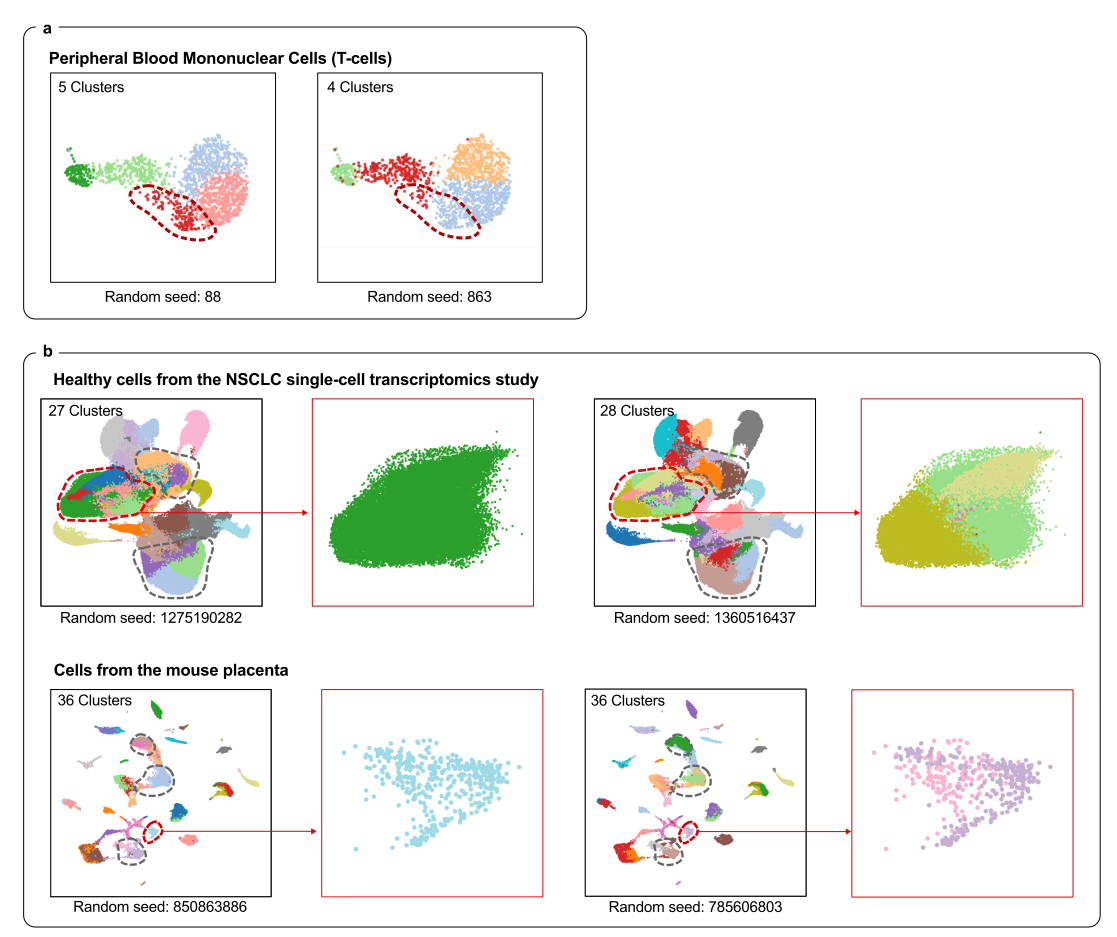
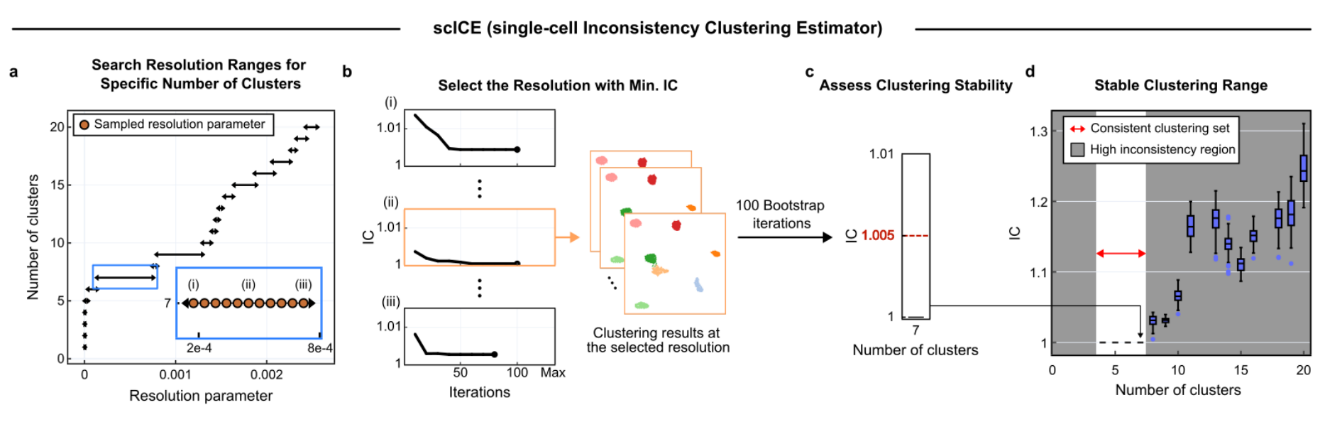
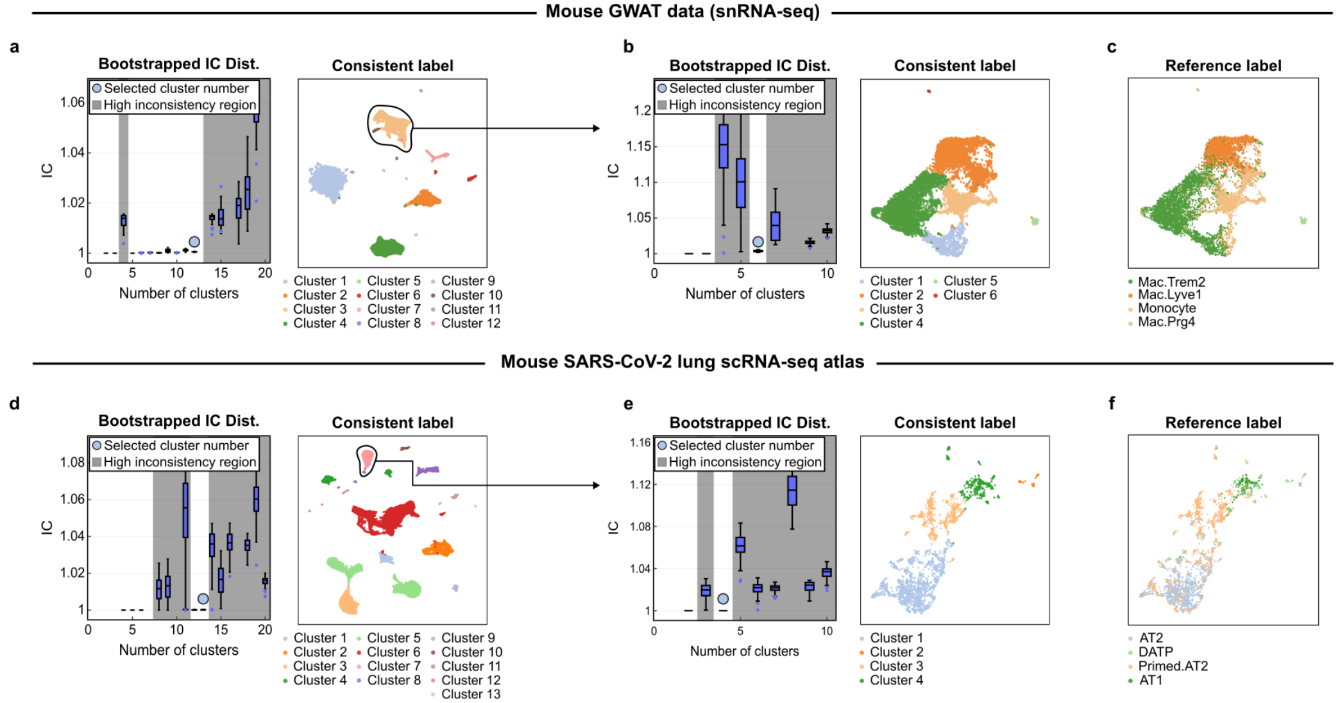
Scientists Tackle Single-Cell Data’s “Reliability Crisis” with New Tool ‘scICE’– scICE validates clustering results up to 30× faster, resolving instability in one of biology’s most widely used analysis methods – The ability to analyze gene expression at the single-cell level — known as single-cell RNA sequencing (scRNA-seq) — has transformed life sciences, driving discoveries across immunology, oncology, and developmental biology. Over 40,000 studies have leveraged this technique to map the complex diversity of cells within tissues and organisms. Yet beneath this explosive growth lies a persistent problem: clustering instability. When researchers attempt to group cells by expression patterns to identify cell types or disease states, they often face inconsistent results — even when analyzing the same dataset repeatedly. Inaccurate clustering can lead to misclassifying normal cells as cancerous or missing rare but critical cell types — jeopardizing interpretation and therapeutic decisions. This “reliability crisis” forces scientists to rerun analyses or rely on computationally expensive pipelines to extract trustworthy insights. Now, a research team led by Professor KIM Jae Kyoung of the Korea Advanced Institute of Science and Technology (KAIST) and the Institute for Basic Science (IBS) has developed a solution: a mathematical framework named scICE (single-cell Inconsistency Clustering Estimator). Traditionally, clustering reliability is assessed by deriving a consensus through repeated analysis of whether individual cell pairs are classified into the same cluster. However, this approach is a computationally demanding process, ill-suited for large-scale datasets with tens of thousands of cells. In contrast, scICE can be applied to large-scale datasets as it bypasses the computationally demanding process of pairwise co-clustering. It instead employs a mathematically defined Inconsistency Coefficient (IC) to assess the stability of cell assignments directly. This allows the tool to efficiently detect and filter out unreliable assignments, preserving only the most stable and biologically meaningful clusters. Dr. KIM Hyun, the first author of the paper (IBS), explained, “Reliability in single-cell clustering has long been overlooked. scICE opens a new path for quickly and easily verifying results.” The research team validated the effectiveness of scICE by applying it to 48 real and simulated scRNA-seq datasets collected from various tissues, including the brain, lungs, and blood. The results revealed that approximately two-thirds of existing analyses were statistically unstable and unreliable. Meanwhile, scICE efficiently selected only a small number of reliable results, saving researchers' time and computational resources while maintaining high accuracy. scICE provides a way to validate clustering outcomes mathematically, ensuring higher confidence in conclusions drawn from single-cell data. Additionally, scICE has drawn attention for its ability to effectively detect rare cell types, which are often overlooked by conventional clustering methods. In practice, scICE reliably identified rare immune cells that can be easily missed in conventional analyses, using subclustering based on its framework. Corresponding author Professor KIM Jae Kyoung stated, “scICE will help researchers swiftly pursue follow-up studies based on reliable results. I hope it becomes a standard tool for trustworthy data interpretation across the life sciences.” The research team made scICE publicly available on [GitHub](https://github.com/Mathbiomed/scICE) for easy access by all users.
Notes for editors
- References
- Media Contact
- About the Institute for Basic Science (IBS) |
| before |
|---|
- Content Manager
- Public Relations Team : Yim Ji Yeob 042-878-8173
- Last Update 2023-11-28 14:20