주메뉴
- About IBS 연구원소개
-
Research Centers
연구단소개
- Research Outcomes
- Mathematics
- Physics
- Center for Underground Physics
- Center for Theoretical Physics of the Universe (Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe (Cosmology, Gravity and Astroparticle Physics Group)
- Dark Matter Axion Group
- Center for Artificial Low Dimensional Electronic Systems
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Exotic Nuclear Studies
- Center for Van der Waals Quantum Solids
- Center for Relativistic Laser Science
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research (Neuro Technology Group)
- Center for Neuroscience Imaging Research (Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Genome Engineering
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Center for Quantum Conversion Research
- Institutes
- Korea Virus Research Institute
- News Center 뉴스 센터
- Career 인재초빙
- Living in Korea IBS School-UST
- IBS School 윤리경영


주메뉴
- About IBS
-
Research Centers
- Research Outcomes
- Mathematics
- Physics
- Center for Underground Physics
- Center for Theoretical Physics of the Universe (Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe (Cosmology, Gravity and Astroparticle Physics Group)
- Dark Matter Axion Group
- Center for Artificial Low Dimensional Electronic Systems
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Exotic Nuclear Studies
- Center for Van der Waals Quantum Solids
- Center for Relativistic Laser Science
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research (Neuro Technology Group)
- Center for Neuroscience Imaging Research (Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Genome Engineering
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Center for Quantum Conversion Research
- Institutes
- Korea Virus Research Institute
- News Center
- Career
- Living in Korea
- IBS School
News Center
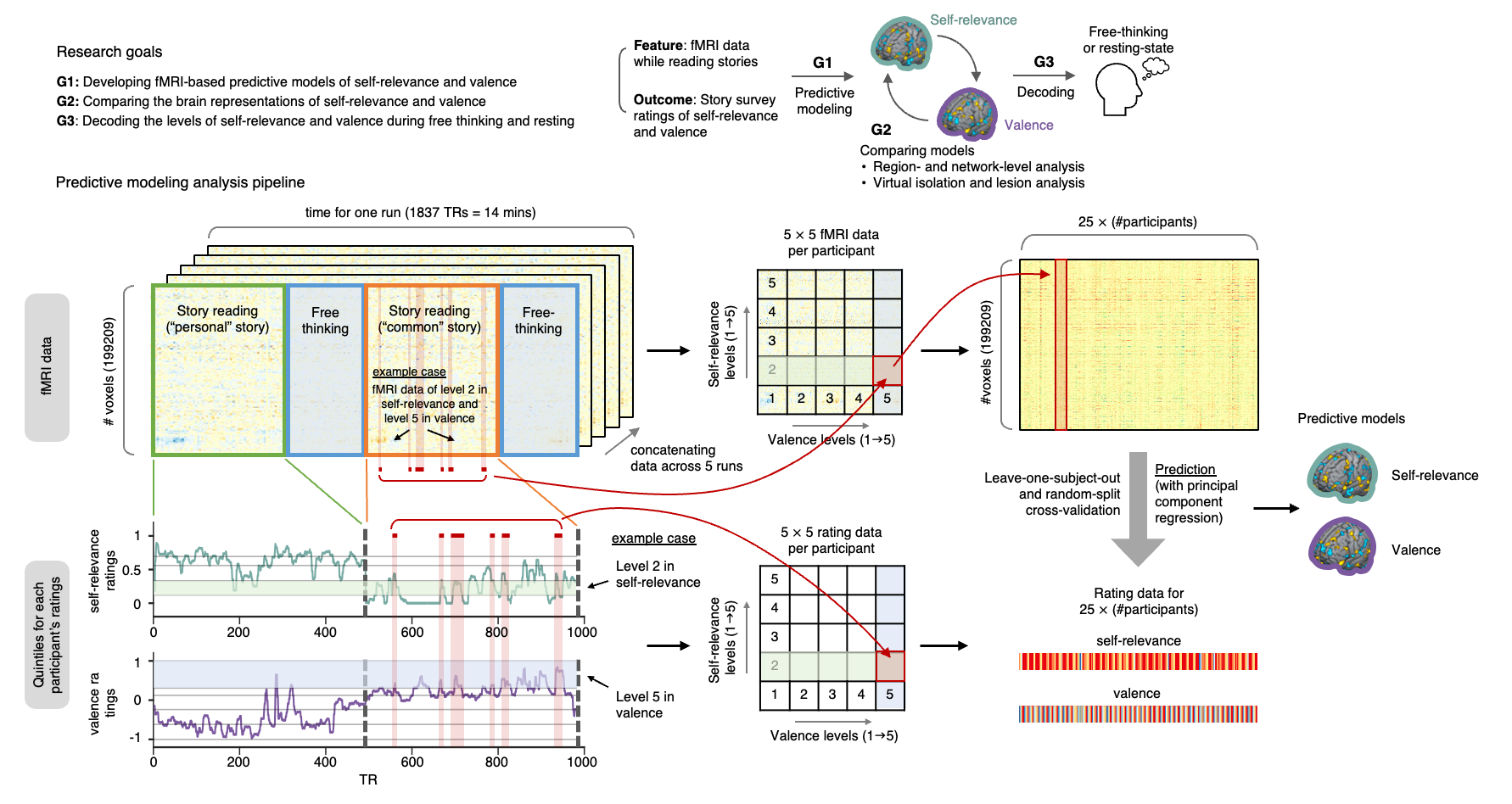
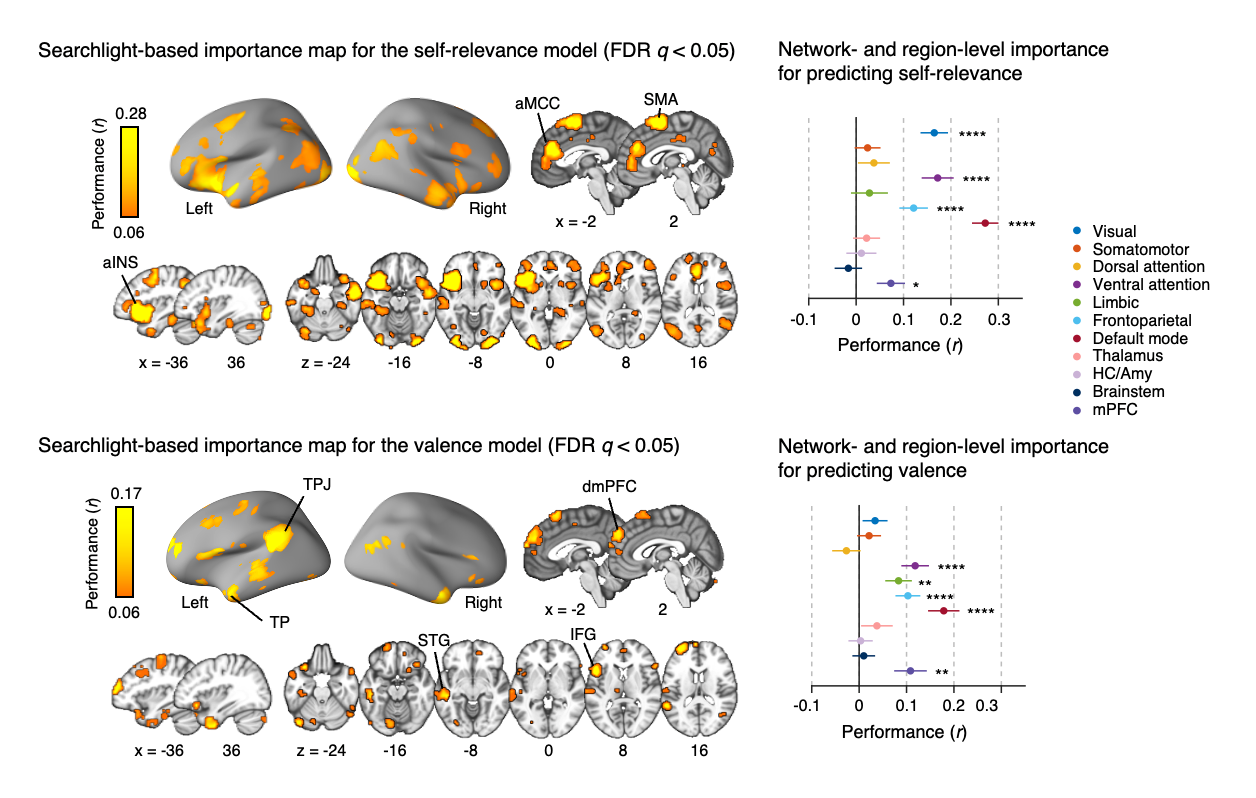
Decoding Spontaneous Thoughts from the Brain via Machine Learning- Predicting self-relevance and valence during personal narratives using fMRI and predictive modeling - A team of researchers led by KIM Hong Ji and WOO Choong-Wan at the Center for Neuroscience Imaging Research (CNIR) within the Institute for Basic Science (IBS), in collaboration with Emily FINN at Dartmouth College, has unlocked a new realm of understanding within the human brain. The team demonstrated the possibility of using functional Magnetic Resonance Imaging (fMRI) and machine learning algorithms to predict subjective feelings in people’s thoughts while reading stories or in a freely thinking state. The brain is constantly active, and spontaneous thoughts occur even during rest or sleep. These thoughts can be anything ranging from memories of the past to aspirations for the future, and they are often intertwined with emotions and personal concerns. However, because spontaneous thought typically occurs without any constraint of consciousness, researching them poses challenges—even simply asking individuals what they are currently thinking can change the nature of their thoughts. New research suggests that it may be possible to develop predictive models of affective contents during spontaneous thought by combining personal narratives with fMRI. Narratives and spontaneous thoughts share similar characteristics, including rich semantic information and temporally unfolding nature. To capture a diverse range of thought patterns, participants engaged in one-on-one interviews to craft personalized narrative stimuli, reflecting their past experiences and emotions. While participants read their stories inside the MRI scanner, their brain activity was recorded. After the fMRI scan, the participants were asked to read the stories again and report perceived self-relevance (i.e., how much this content is related to themselves) and valence (i.e., how much this content is positive or negative) at each moment. Using a quintile (five levels) from each participant's self-relevance and valence ratings, 25 (5 levels of self-relevance rating × 5 levels of valence rating) possible segments of fMRI and rating data were created. The team then harnessed machine learning techniques to train predictive models, combining these data with the fMRI brain scans from 49 individuals to decode the “emotional dimensions” of thoughts in real time. To interpret the brain representations of the predictive models, the research team employed multiple approaches, such as virtual lesion and virtual isolation analyses at both region and network levels. Through these analyses, they discovered the significance of the default mode, ventral attention, and frontoparietal networks in both self-relevance and valence predictions. Specifically, they identified the involvement of the anterior insula and midcingulate cortex in self-relevance prediction, while the left temporoparietal junction and dorsomedial prefrontal cortex played important roles in valence prediction. Moreover, the predictive models showed their capacity to predict both self-relevance and valence not only during story reading but also when applied to data from 199 individuals engaging in spontaneous, task-free thinking or even during resting. These findings show the promise of daydream decoding. “Several tech companies and research teams are currently endeavoring to decode words or images directly from brain activity, but there are limited initiatives aimed at decoding intimate emotions underlying these thoughts,” stated Dr. WOO Choong-Wan, associate director of IBS, who led the study. “Our research is centered on human emotions, with the aim of decoding emotions within the natural flow of thoughts to obtain information that can benefit people’s mental health.” KIM Hongji, a doctoral candidate and the first author of this study, emphasized, "This study holds significance as we decoded the emotional state associated with general thoughts, rather than targeting emotions limited to specific tasks," adding, "These findings advance our understanding of the internal states and contexts influencing subjective experiences, potentially shedding light on individual differences in thoughts and emotions, and aiding in the evaluation of mental well-being."
Notes for editors
- References
- Video abstract
- Media Contact
- About the Institute for Basic Science (IBS)
|
| Next | |
|---|---|
| before |
- Content Manager
- Public Relations Team : Yim Ji Yeob 042-878-8173
- Last Update 2023-11-28 14:20