주메뉴
- About IBS 연구원소개
-
Research Centers
연구단소개
- Research Outcomes
- Mathematics
- Physics
- Center for Underground Physics
- Center for Theoretical Physics of the Universe (Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe (Cosmology, Gravity and Astroparticle Physics Group)
- Dark Matter Axion Group
- Center for Artificial Low Dimensional Electronic Systems
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Exotic Nuclear Studies
- Center for Van der Waals Quantum Solids
- Center for Relativistic Laser Science
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research (Neuro Technology Group)
- Center for Neuroscience Imaging Research (Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Institutes
- Korea Virus Research Institute
- News Center 뉴스 센터
- Career 인재초빙
- Living in Korea IBS School-UST
- IBS School 윤리경영


주메뉴
- About IBS
-
Research Centers
- Research Outcomes
- Mathematics
- Physics
- Center for Underground Physics
- Center for Theoretical Physics of the Universe (Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe (Cosmology, Gravity and Astroparticle Physics Group)
- Dark Matter Axion Group
- Center for Artificial Low Dimensional Electronic Systems
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Exotic Nuclear Studies
- Center for Van der Waals Quantum Solids
- Center for Relativistic Laser Science
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research (Neuro Technology Group)
- Center for Neuroscience Imaging Research (Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Institutes
- Korea Virus Research Institute
- News Center
- Career
- Living in Korea
- IBS School
News Center
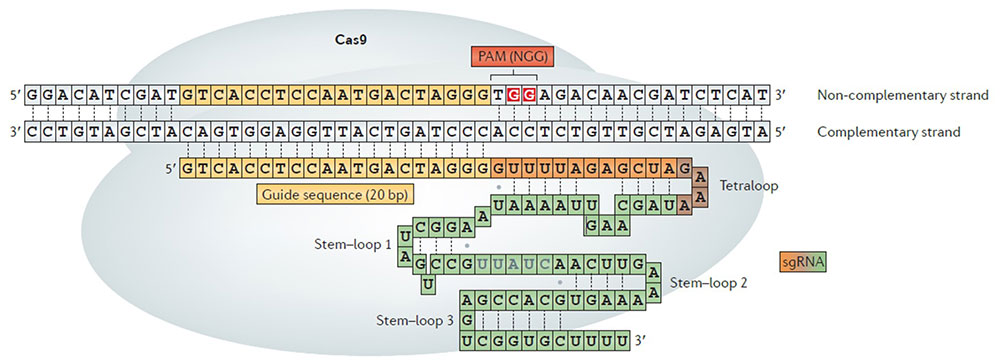
Comparing 13 different CRISPR-Cas9 DNA scissorsComputational models based on deep learning predict the activities of SpCas9 variants and provide a useful guide for selecting the most appropriate one CRISPR-Cas9 has become one of the most convenient and effective biotechnology tools used to cut specific DNA sequences. Starting from Streptococcus pyogenes Cas9 (SpCas9), a multitude of variants have been engineered and employed for experiments worldwide. Although all these systems are targeting and cleaving a specific DNA sequence, they also exhibit relatively high off-target activities with potentially harmful effects. Led by Professor Hyongbum Henry Kim, the research team of the Center for Nanomedicine, within the Institute for Basic Science (IBS, South Korea), has achieved the most extensive high-throughput analysis of CRISPR-Cas9 activities. The team developed deep-learning-based computational models that predict the activities of SpCas9 variants for different DNA sequences. Published in Nature Biotechnology, this study represents a useful guide for selecting the most appropriate SpCas9 variant.
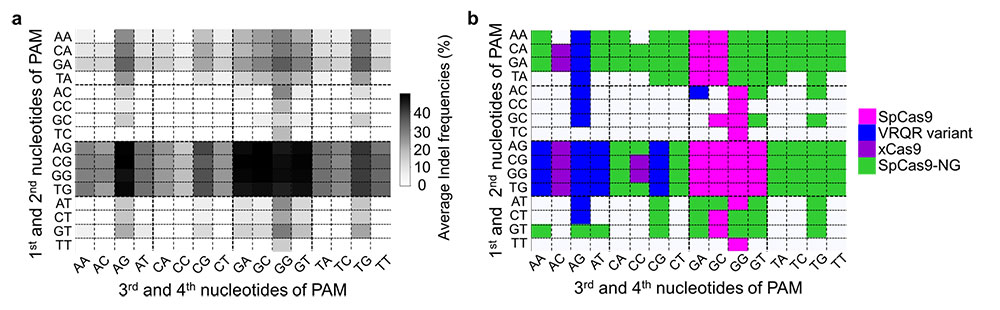
This study surpassed all previous reports, which had evaluated only up to three Cas9 systems. IBS researchers compared 13 SpCas9 variants and defined which four-nucleotide sequences can be used as protospacer adjacent motif (PAM) – a short DNA sequence that is required for Cas9 to cut and is positioned immediately after the DNA sequence targeted for cleavage.
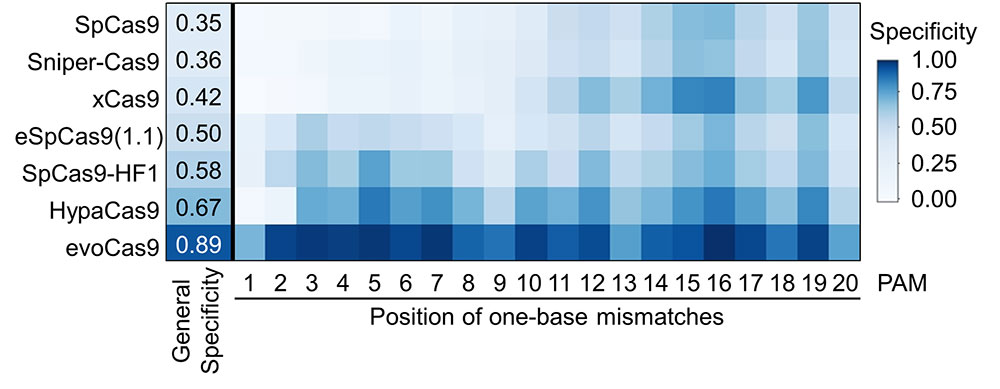
Additionally, they evaluated the specificity of six different high-fidelity SpCas9 variants, and found that evoCas9 has the highest specificity, while the original wild-type SpCas9 has the lowest. Although evoCas9 is very specific, it also shows low activity at many target sequences: these results imply that, depending on the DNA target sequence, other high-fidelity Cas9 variants could be preferred.
Based on these results, IBS researchers developed DeepSpCas9variants (http://deepcrispr.info/DeepSpCas9variants/), a computational tool to predict the activities of SpCas9 variants. By accessing this public website, users may input the desired DNA target sequence, find out the most suitable SpCas9 variant and take full advantage of the CRISPR technology. “We began this research when we noticed the critical lack of a systematic comparison among the different SpCas9 variants,” says Kim. “Now, using DeepSpCas9variants, researchers can select the most appropriate SpCas9 variants for their own research purposes.” Notes for editors - References - Media Contact - About the Institute for Basic Science (IBS) |
|||
|
|
| Next | |
|---|---|
| before |
- Content Manager
- Public Relations Team : Yim Ji Yeob 042-878-8173
- Last Update 2023-11-28 14:20