주메뉴
- About IBS 연구원소개
-
Research Centers
연구단소개
- Research Outcomes
- Mathematics
- Physics
- Center for Underground Physics
- Center for Theoretical Physics of the Universe (Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe (Cosmology, Gravity and Astroparticle Physics Group)
- Dark Matter Axion Group
- Center for Artificial Low Dimensional Electronic Systems
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Exotic Nuclear Studies
- Center for Van der Waals Quantum Solids
- Center for Relativistic Laser Science
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research (Neuro Technology Group)
- Center for Neuroscience Imaging Research (Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Genome Engineering
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Institutes
- Korea Virus Research Institute
- News Center 뉴스 센터
- Career 인재초빙
- Living in Korea IBS School-UST
- IBS School 윤리경영


주메뉴
- About IBS
-
Research Centers
- Research Outcomes
- Mathematics
- Physics
- Center for Underground Physics
- Center for Theoretical Physics of the Universe (Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe (Cosmology, Gravity and Astroparticle Physics Group)
- Dark Matter Axion Group
- Center for Artificial Low Dimensional Electronic Systems
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Exotic Nuclear Studies
- Center for Van der Waals Quantum Solids
- Center for Relativistic Laser Science
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research (Neuro Technology Group)
- Center for Neuroscience Imaging Research (Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Genome Engineering
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Institutes
- Korea Virus Research Institute
- News Center
- Career
- Living in Korea
- IBS School
News Center
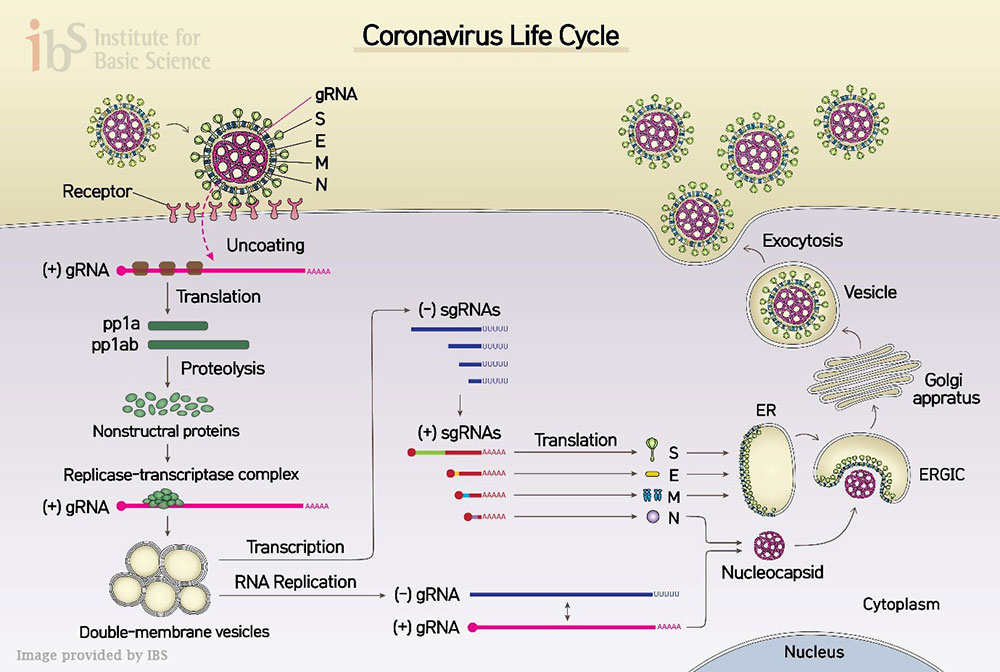
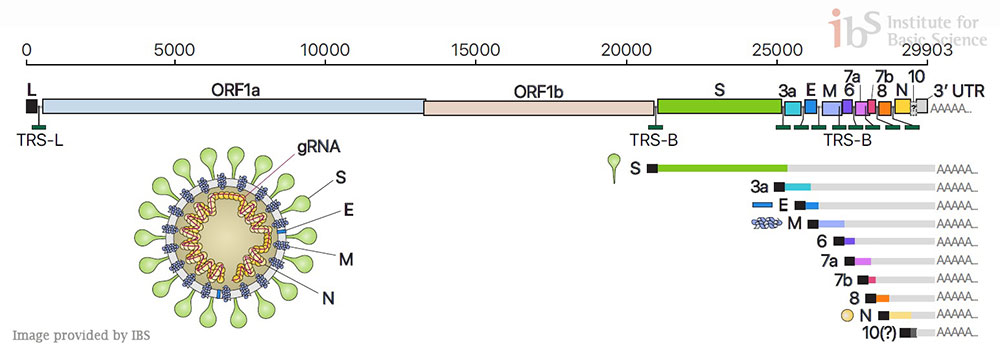
New Coronavirus (SARS-CoV-2) Mapped OutA high resolution gene map reveals many viral RNAs with unknown functions and modifications Jean and Peter Medawar wrote in 1977 that a virus is “simply a piece of bad news wrapped up in proteins.” The “bad news” in the SARS-CoV-2 case is the genome made of a very long ribonucleic acid (RNA) molecule. Grappling with COVID-19 pandemic, the world seems to be lost with no sense of direction in uncovering what this coronavirus (SARS-Cov-2) is composed of. Being an RNA virus, SARS-Cov-2 enters host cells and replicates its genomic RNA and produces many smaller RNAs (called “subgenomic RNAs”). These subgenomic RNAs are used for the synthesis of various proteins (spikes, envelopes, etc.) that are required for the beginning of SARS-Cov-2 lineage. Thus, the smaller RNAs make good targets for messing up new coronavirus’s conquering of our immune system. Though recent studies reported the sequence of the RNA genome, they only predicted where their genes might be, without concretely pinpointing what kind of genes are present and where exactly in the genome.
Led by Professors KIM Narry and CHANG Hyeshik, the Center for RNA Research within the Institute for Basic Science (IBS), South Korea, succeeded in dissecting the architecture of SARS-CoV-2 RNA genome, in collaboration with Korea Centers for Disease Control & Prevention (KCDC). The researchers experimentally confirmed the predicted subgenomic RNAs that are in turn translated into viral proteins. Furthermore, they analyzed the sequence information of each RNA and revealed where genes are exactly located on a genomic RNA. “Not only detailing the gene structure of SARS-CoV-2, we also discovered numerous new RNAs. Our work provides a high-resolution map of SARS-CoV-2. This map will help understand how the virus replicates and how it escapes the human defense system,” explains Professor KIM Narry, one of the corresponding authors of the study. It was previously predicted that 10 subgenomic RNAs make up the viral particle structure. However, the research team confirmed that 9 subgenomic RNAs actually exist, invalidating one. Researchers also found that there are dozens of unknown subgenomic RNAs, owing to RNA fusion and deletion events. “Though it requires further investigation, these molecular events may lead to the relatively rapid evolution of coronavirus. It is unclear yet what these novel RNAs do, but a possibility is that they may assist the virus to avoid the attack from the host,” says Prof. Kim.
They believe if they figure out the unknown characteristics of RNA, the findings may offer a new clue for combatting the new coronavirus. Newly discovered features will also help to understand the life cycle of the virus and develop new strategies for antiviral therapy. Behind the success of the study is the research team’s pairing of two complementary sequencing techniques; DNA nanoball sequencing and nanopore direct RNA sequencing. The nanopore direct RNA sequencing allows to directly analyze the entire long viral RNA without fragmentation. Conventional RNA sequencing methods usually require a step-by-step process of cutting and converting RNA to DNA before reading RNA. Meanwhile, the DNA nanoball sequencing can read only short fragments, but has the advantage of analyzing a large number of sequences with high accuracy. These two techniques turned out to be highly complementary to each other to analyze the viral RNAs. “Now we have secured a high resolution gene map of the new coronavirus that guides us where to find each bit of genes on all of the total SARS-CoV-2 RNAs (transcriptome) and all modifications RNAs (epitranscriptome). It is time to explore the functions of the newly discovered genes and the mechanism underlying viral gene fusion. We believe that our study will contribute to the development of diagnostics and therapeutics to combat the virus more effectively,” notes Professor KIM Narry. Dahee Carol Kim Notes for editors - References - Media Contact - About the Institute for Basic Science (IBS) |
|||
|
|
| Next | |
|---|---|
| before |
- Content Manager
- Public Relations Team : Yim Ji Yeob 042-878-8173
- Last Update 2023-11-28 14:20