주메뉴
- About IBS 연구원소개
-
Research Centers
연구단소개
- Research Outcomes
- Mathematics
- Physics
- Center for Theoretical Physics of the Universe(Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe(Cosmology, Gravity and Astroparticle Physics Group)
- Center for Exotic Nuclear Studies
- Center for Artificial Low Dimensional Electronic Systems
- Center for Underground Physics
- Center for Axion and Precision Physics Research
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Van der Waals Quantum Solids
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Institutes
- Korea Virus Research Institute
- News Center 뉴스 센터
- Career 인재초빙
- Living in Korea IBS School-UST
- IBS School 윤리경영


주메뉴
- About IBS
-
Research Centers
- Research Outcomes
- Mathematics
- Physics
- Center for Theoretical Physics of the Universe(Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe(Cosmology, Gravity and Astroparticle Physics Group)
- Center for Exotic Nuclear Studies
- Center for Artificial Low Dimensional Electronic Systems
- Center for Underground Physics
- Center for Axion and Precision Physics Research
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Van der Waals Quantum Solids
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Institutes
- Korea Virus Research Institute
- News Center
- Career
- Living in Korea
- IBS School
News Center
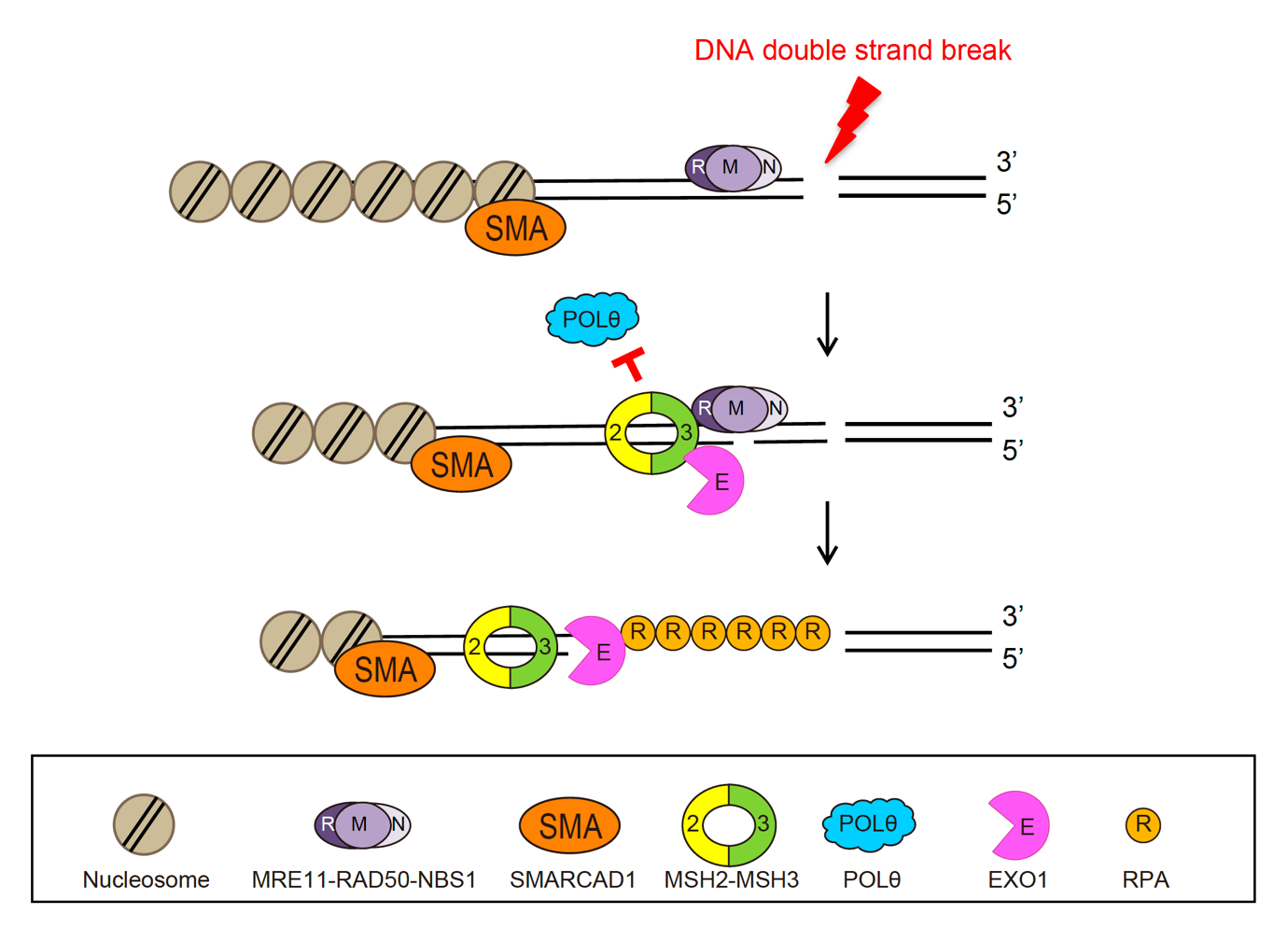
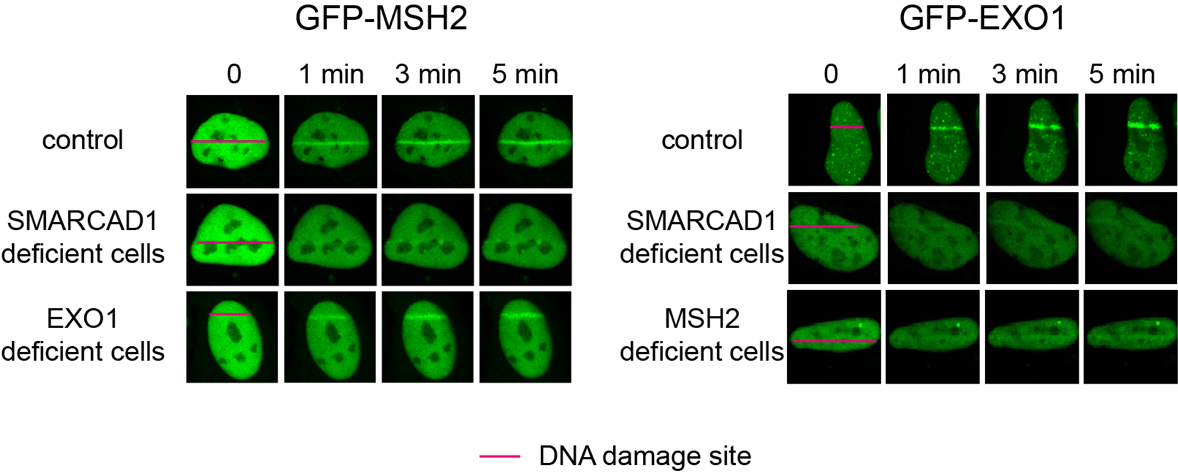
How cells select DNA damage repair pathwaysA new function of mismatch repair protein MSH2-MSH3 in DNA double-strand break repair DNA is well known as the blueprint of life, necessary for an organism to facilitate living processes. DNA can be damaged by various factors such as radical metabolites, radiation, and some toxic chemicals. As DNA is a molecule consisting of two strands, either one or both of the strands can be damaged. Single-strand break (SSB), occur when one of the two strands of DNA is damaged or broken. These are relatively mild damages that can be easily repaired by specialized enzymes that can seal the break and restore the integrity of the DNA molecule. On the other hand, a double-strand break (DSB) refers to when both strands of DNA are damaged. These are considered the most severe type of DNA damage, capable of causing genetic mutations or cell death. Cells maintain genome integrity by having various pathways to repair DSBs. Among the several mechanisms for repairing DSBs, homologous recombination repair (HR) is one such mechanism that is highly precise and error-free, as it uses the undamaged sister chromatid as a template to repair DSBs. On the other hand, DNA repaired by polymerase theta-mediated end-joining (TMEJ) can result in the loss of some genetic information and cause mutations. Therefore, it is crucial to choose the appropriate DSB repair process to maintain genome integrity. But How do cells select the right repair process? And what kinds of proteins are involved in the selection process? Led by Professor MYUNG Kyungjae, Director of the Center of Genomic Integrity (CGI) within the Institute for Basic Science (IBS), the research teams of Professor LEE Ja Yil at Ulsan National Institute of Science and Technology, and Professor OH Jung-Min at Pusan National University have discovered that repair proteins involved in DSB repair, mismatch repair, and TMEJ are closely related and interact with each other during DSB repair process. There are various repair mechanisms in our cells, each tailored to the type of DNA damage. For instance, DSBs are repaired by DSB repair proteins, while improperly paired DNA bases are repaired by mismatch repair proteins. Until now, most researchers thought that a specific type of DNA damage is only repaired by its corresponding DNA repair mechanism. However, this study revealed that repair proteins previously thought to be responsible for different repair mechanisms can interact with each other to recognize damaged sites and select an appropriate repair mechanism. Specifically, it was revealed that MSH2-MSH3, a DNA mismatch repair protein, actually plays a crucial role in the DSB repair process. The researchers observed the recruitment of fluorescent protein-labeled MSH2-MSH3 protein to the site of DSBs and revealed that this movement occurs through binding with the chromatin remodeling protein called SMARCAD1. The binding of MSH2-MSH3 to DSBs facilitates the recruitment of EXO1 (exonuclease 1) for long-range resection of damaged DNA. After the long-range resection, the damaged DNA is repaired through error-free HR. Furthermore, it was discovered that the binding of MSH2-MSH3 inhibits the access of POLθ, which mediates a more error-prone TMEJ pathway, thereby preventing mutations that may occur during DSB repair. Director Myung said, “This research has revealed a new function of the mismatch repair protein MSH2-MSH3 in regulating DSB repair,” adding, “The repair proteins that have been believed so far to act independently in the mismatch repair, double-stranded break repair, and TMEJ repair pathways are now shown to closely interact each other for proper maintenance of genomic integrity.” This work was published in Nucleic Acids Research on May 4th, 2023.
Notes for editors
- Reference
- Media Contact
- About the Institute for Basic Science (IBS)
|
| Next | |
|---|---|
| before |
- Content Manager
- Public Relations Team : Suh, William Insang 042-878-8137
- Last Update 2023-11-28 14:20