주메뉴
- About IBS 연구원소개
-
Research Centers
연구단소개
- Research Outcomes
- Mathematics
- Physics
- Center for Theoretical Physics of the Universe(Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe(Cosmology, Gravity and Astroparticle Physics Group)
- Center for Exotic Nuclear Studies
- Center for Artificial Low Dimensional Electronic Systems
- Center for Underground Physics
- Center for Axion and Precision Physics Research
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Van der Waals Quantum Solids
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Institutes
- Korea Virus Research Institute
- News Center 뉴스 센터
- Career 인재초빙
- Living in Korea IBS School-UST
- IBS School 윤리경영


주메뉴
- About IBS
-
Research Centers
- Research Outcomes
- Mathematics
- Physics
- Center for Theoretical Physics of the Universe(Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe(Cosmology, Gravity and Astroparticle Physics Group)
- Center for Exotic Nuclear Studies
- Center for Artificial Low Dimensional Electronic Systems
- Center for Underground Physics
- Center for Axion and Precision Physics Research
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Van der Waals Quantum Solids
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Institutes
- Korea Virus Research Institute
- News Center
- Career
- Living in Korea
- IBS School
News Center
Scientists improve the equation in FDA guidance predicting drug interactions- Collaboration of mathematicians and pharmaceutical scientists reveals the fundamental limit of the equation in FDA guidance for drug-drug interaction prediction and derives a dramatically improved equation - Release Summary Text (75 words maximum)The collaboration of mathematicians and pharmaceutical scientists identifies the major cause of the low accuracy of the equation in the FDA guidance, predicting drug-drug interaction through CYP enzyme induction. To overcome the fundamental limit of the conventional equation, the scientists derived a new equation based on mathematical theory. This dramatically improves the accuracy of drug-drug interaction prediction, which is critical to preventing drug failure and enhancing new drug development. Full text of release:Drugs absorbed into the body are metabolized and thus removed by enzymes in several organs like the liver. The clearance rates of the drugs can be increased by other drugs that increase the level of enzymes in the body. This dramatically decreases the concentration of a drug, reducing its efficacy and often leading to the failure of the drug. Therefore, accurately predicting the clearance rate in the presence of drug-drug interaction* is critical in new drug development and prescription. *Drug-drug interaction: In terms of metabolism, drug-drug interaction is a phenomenon in which, when two or more drugs are taken together, one drug changes the metabolism of another drug to promote or inhibit its excretion from the body. As a result, it increases the toxicity of medicines or causes loss of efficacy. To indirectly evaluate drug-drug interaction, pharmaceutical scientists have relied on the 110-year-old Michaelis-Menten (MM) model, which describes the reaction rate of enzymes. Although the MM equation has been one of the most widely known equations in biochemistry (used in more than 220,000 published papers), it has a fundamental limit. That is, the MM equation is accurate only when the concentration of the enzyme that metabolizes the drug is much lower than its MM constant (Km). Furthermore, when enzyme concentration is increased by drug-drug interaction, the MM equation is expected to be extremely inaccurate. Notably, the Food and Drug Administration published the FDA guidance* in 2020, which includes an equation based on this model to predict the change in drug clearance. However, the accuracy of the equation has been highly unsatisfactory – only 38 percent of the predictions had less than two-fold errors. To resolve this, scientifically-unjustified artificial constants have been incorporated into the equation to hopefully improve its prediction. Professor CHAE Jung-woo said, “This is comparable to epicyclic orbits having to be introduced to explain the motion of the planets using the now-defunct Ptolemaic theory.” *Drug-drug interaction is a key mechanism that causes drug toxicity and/or its loss of efficacy. The US FDA has issued guidance to evaluate and prevent drug-drug interactions since the mid-1990s and was revised in 2020 with the latest guidance. A joint research team composed of mathematicians from the Biomedical Mathematics Group within the Institute for Basic Science (IBS) and the Korea Advanced Institute of Science and Technology (KAIST) and pharmacological scientists from the Chungnam National University reported that they identified the major causes of the FDA-recommended equation’s inaccuracies and presented solutions. When estimating the gut bioavailability (Fg), which is the key parameter of the equation, the fraction absorbed from the gut lumen (Fa) is usually assumed to be one. However, many experiments have shown that Fa is less than one, obviously since it can’t be expected that all of the orally taken drugs to be completely absorbed by the intestines. To solve this problem, the research team used an “estimated Fa” value based on factors such as the drug’s transit time, intestine radius, and permeability values and used it to re-calculate Fg. Also, the team used an alternative model they derived in a previous study back in 2020, which can more accurately predict the drug metabolism rate regardless of the enzyme concentration, unlike the MM equation. Combining these changes, the modified equation with re-calculated Fg had a dramatically increased accuracy of about 80%, up from 38%. “Such drastic improvement in drug-drug interaction prediction accuracy is expected to greatly contribute to increasing the success rate of new drug development and drug efficacy in clinical practice. As the results of this study were published in the top clinical pharmacology journal, it is expected that the FDA guidance will be revised according to the results of this study.” said professor KIM Jae Kyoung from the IBS Biomedical Mathematics Group. Furthermore, this study highlights the importance of collaborative research between research groups in vastly different disciplines, in a field that is as dynamic as drug interactions. “Collaboration between various disciplines, especially convergence research with mathematics, which is a basic science, reminded me of a proverb; going alone goes fast, but going together goes far,” said professor KIM Sang Kyum.
Notes for editors
- Reference
- Media Contact
- About the Institute for Basic Science (IBS)
|
| Next | |
|---|---|
| before |
- Content Manager
- Public Relations Team : Suh, William Insang 042-878-8137
- Last Update 2023-11-28 14:20










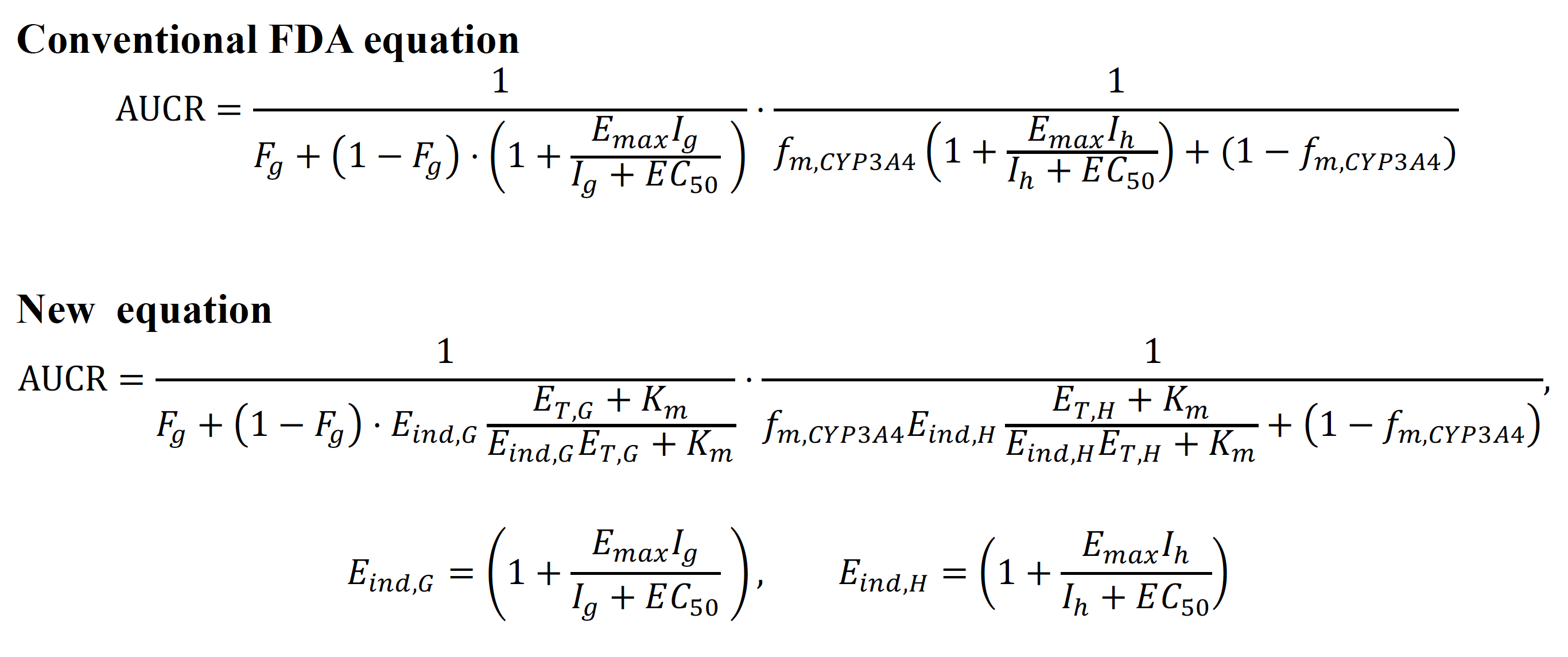 Figure 1. The conventional equation recommended by the FDA guidance (upper) and the newly derived equation (lower) for predicting drug-drug interaction. The FDA guidance has recommended the upper equation for predicting the drug-drug interaction mediated by enzyme induction. However, due to its low accuracy, artificial constants have been used to calibrate the equation. To address this issue, the scientists identified the major cause of the inaccuracy of the conventional equation and derived a new equation based on rigorous mathematical theory.
Figure 1. The conventional equation recommended by the FDA guidance (upper) and the newly derived equation (lower) for predicting drug-drug interaction. The FDA guidance has recommended the upper equation for predicting the drug-drug interaction mediated by enzyme induction. However, due to its low accuracy, artificial constants have been used to calibrate the equation. To address this issue, the scientists identified the major cause of the inaccuracy of the conventional equation and derived a new equation based on rigorous mathematical theory.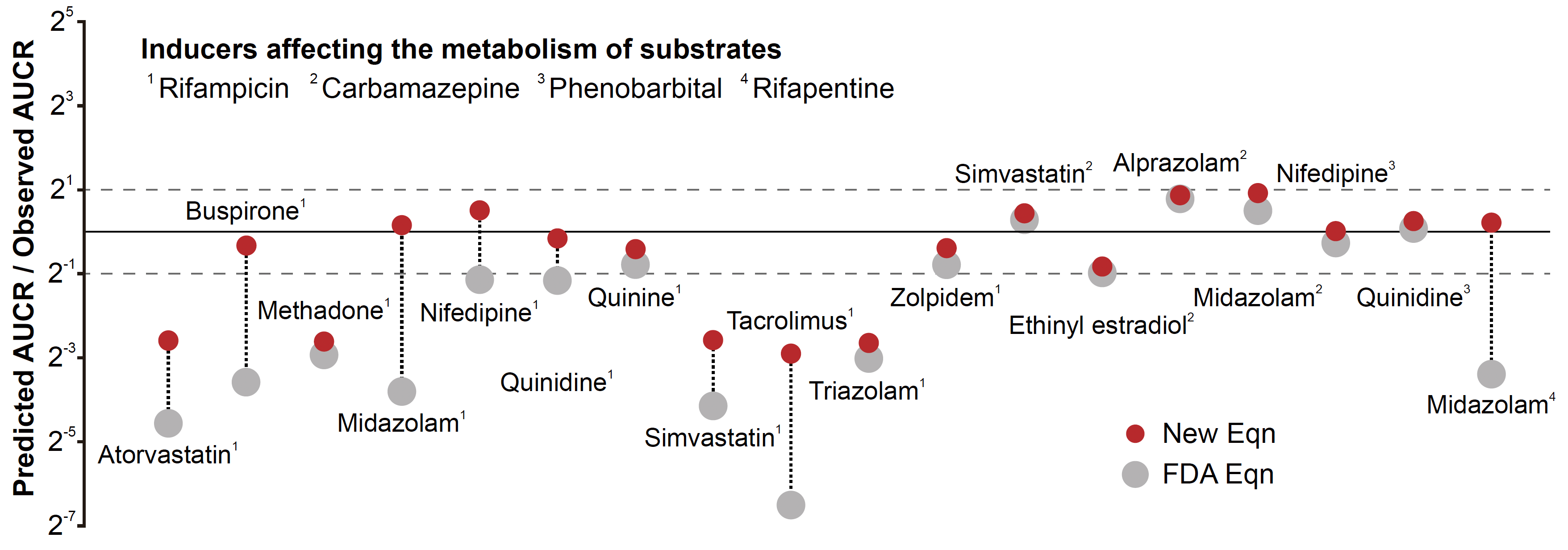 Figure 2. The new equation provides more accurate predictions than the conventional FDA equation. The FDA equation with Fg estimated assuming Fa = 1 underpredicts the area under the curve ratio (AUCR), which represents the drug-drug interaction mediated by CYP enzyme induction, for numerous drug pairs (grey dots). On the other hand, the new equation with re-estimated Fg predicts the AUCR within two-fold errors for nearly double the number of drug pairs than the conventional FDA equation. Solid and dashed lines are the line of precise prediction and two-fold error, respectively.
Figure 2. The new equation provides more accurate predictions than the conventional FDA equation. The FDA equation with Fg estimated assuming Fa = 1 underpredicts the area under the curve ratio (AUCR), which represents the drug-drug interaction mediated by CYP enzyme induction, for numerous drug pairs (grey dots). On the other hand, the new equation with re-estimated Fg predicts the AUCR within two-fold errors for nearly double the number of drug pairs than the conventional FDA equation. Solid and dashed lines are the line of precise prediction and two-fold error, respectively.
