주메뉴
- About IBS 연구원소개
-
Research Centers
연구단소개
- Research Outcomes
- Mathematics
- Physics
- Center for Underground Physics
- Center for Theoretical Physics of the Universe (Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe (Cosmology, Gravity and Astroparticle Physics Group)
- Dark Matter Axion Group
- Center for Artificial Low Dimensional Electronic Systems
- Center for Quantum Nanoscience
- Center for Exotic Nuclear Studies
- Center for Van der Waals Quantum Solids
- Center for Relativistic Laser Science
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research (Neuro Technology Group)
- Center for Neuroscience Imaging Research (Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Institutes
- Korea Virus Research Institute
- News Center 뉴스 센터
- Career 인재초빙
- Living in Korea IBS School-UST
- IBS School 윤리경영


주메뉴
- About IBS
-
Research Centers
- Research Outcomes
- Mathematics
- Physics
- Center for Underground Physics
- Center for Theoretical Physics of the Universe (Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe (Cosmology, Gravity and Astroparticle Physics Group)
- Dark Matter Axion Group
- Center for Artificial Low Dimensional Electronic Systems
- Center for Quantum Nanoscience
- Center for Exotic Nuclear Studies
- Center for Van der Waals Quantum Solids
- Center for Relativistic Laser Science
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research (Neuro Technology Group)
- Center for Neuroscience Imaging Research (Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Institutes
- Korea Virus Research Institute
- News Center
- Career
- Living in Korea
- IBS School
News Center
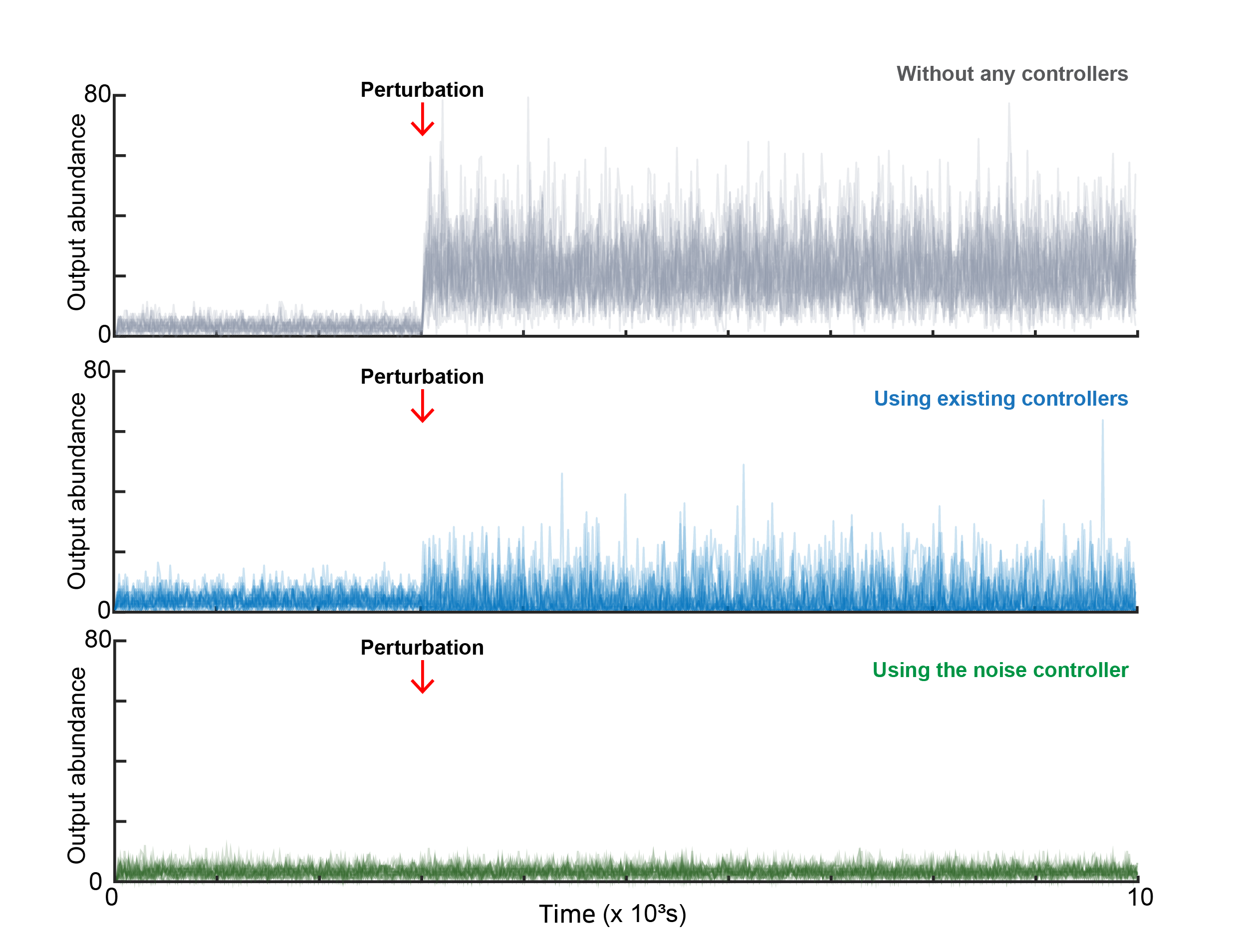
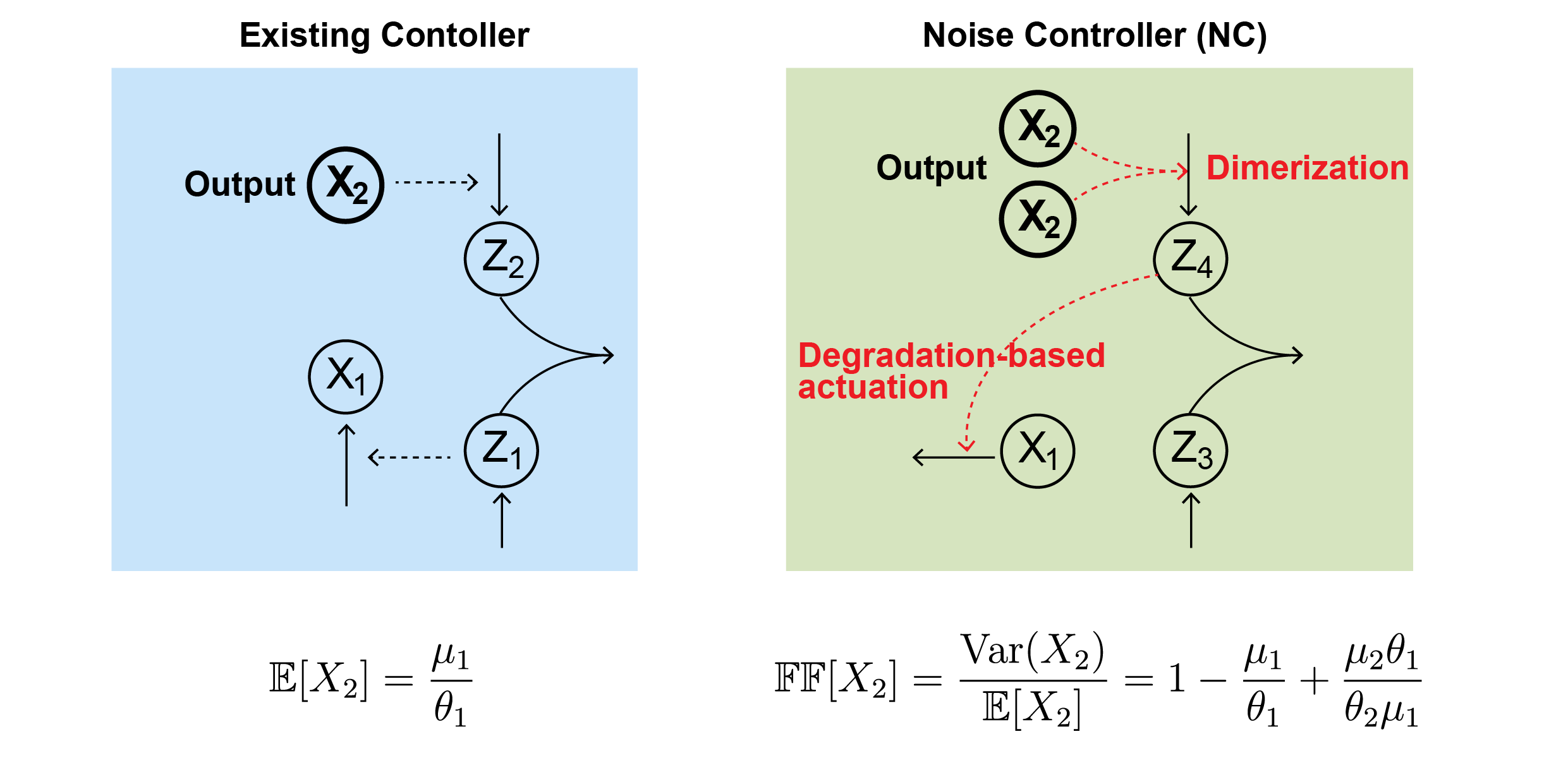
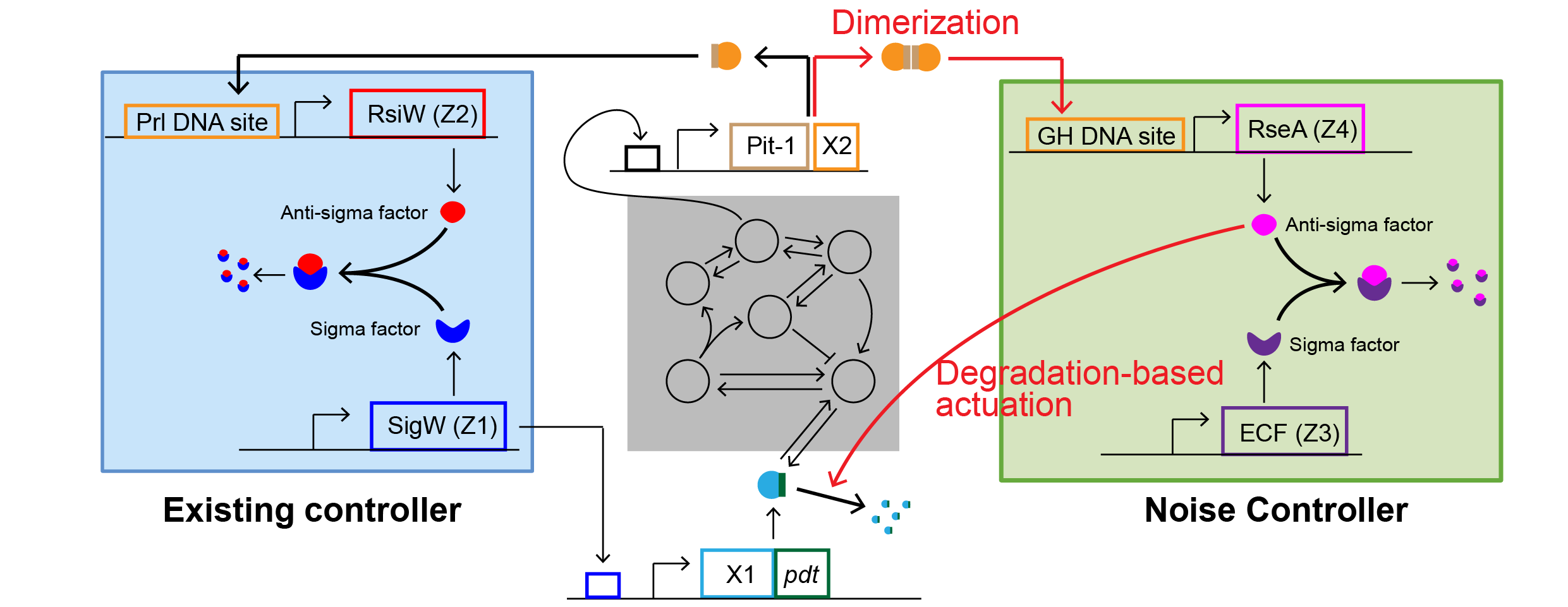
Mathematicians Tame Cellular “Noise” to Control Life at the Single-Cell Level- Researchers develop a “Noise Controller” that solves a decades-old biological puzzle, paving the way for precision cancer therapy and synthetic biology - Why does cancer sometimes recur after chemotherapy? Why do some bacteria survive antibiotic treatment? In many cases, the answer appears to lie not in genetic differences, but in biological noise — random fluctuations in molecular activity that occur even among genetically identical cells. Biological systems are inherently noisy, as molecules inside living cells are produced, degraded, and interact through fundamentally random processes. Understanding how biological systems cope with such fluctuations — and how they might be controlled — has been a long-standing challenge in systems and synthetic biology. Although modern biology can regulate the average behavior of a cell population, controlling the unpredictable fluctuations of individual cells has remained a major challenge. These rare “outlier” cells, driven by stochastic variation, can behave differently from the majority and influence system-level outcomes. This longstanding problem has been answered by a joint research team led by Professor KIM Jae Kyoung (KAIST, IBS Biomedical Mathematics Group), KIM Jinsu (POSTECH), and Professor CHO Byung-Kwan (KAIST), which has developed a novel mathematical framework called the “Noise Controller” (NC). This achievement establishes a level of single-cell precision control previously thought impossible, and it is expected to provide a key breakthrough for longstanding challenges in cancer therapy and synthetic biology. The “Freezing and Boiling Shower” ProblemCells maintain life through homeostasis — keeping internal conditions stable despite external changes. Synthetic biologists have long tried to engineer circuits to control the protein levels within the cells. Traditional strategies in biology are designed to stabilize mean behavior using feedback control. However, regulating averages alone using this route can come at a cost. Previous studies have shown that certain feedback mechanisms can unintentionally amplify variability, making systems more noisy rather than less. This trade-off has been viewed as a fundamental barrier, raising the question of whether precise control of stochastic biological systems is even possible. The research team compares this to adjusting a shower. “Standard control methods are like adjusting a shower,” they explained. “You might get the water to average 40°C, but if that average is achieved by alternating between freezing cold and boiling hot water, you can’t take a shower. Similarly, in biology, getting the average right isn’t enough if individual cells are fluctuating wildly.” This fluctuation is dangerous. In disease treatment, the “outlier” cells — those fluctuating away from the average — are often the ones that develop drug resistance, leading to cancer recurrence or chronic infection. The Mathematical Solution: “Noise Robust Perfect Adaptation”To solve this, the team designed a new gene regulatory circuit using mathematical modeling. The researchers instead pursued an approach that directly targets noise, rather than treating it as an unavoidable side effect. Unlike previous controllers that only sensed protein abundance, the new Noise Controller (NC) creates a feedback loop that senses the “noise” itself, specifically, the second moment of protein levels. The key discovery was a mechanism involving dimerization (where two proteins bind together) combined with degradation-based actuation (actively breaking down specific proteins). This setup allows the cell to effectively “measure” and dampen its own internal noise. The result is a state the researchers call “Noise Robust Perfect Adaptation” (Noise RPA). This technology allows for a regime in which both the average protein level and the magnitude of stochastic fluctuations remain stable, even under changing conditions. Importantly, the model shows that noise can be reduced down to a level commonly regarded as a fundamental physical limit imposed by stochastic molecular processes, characterized by a Fano factor of 1. By introducing this novel control architecture, the team demonstrated that it is possible to suppress stochastic fluctuations while simultaneously maintaining stable average behavior. From Theory to CureThe team validated this technology through rigorous computer simulations (in silico experiments) using the DNA repair system of E. coli. In a standard simulation, approximately 20% of bacteria failed to activate their DNA repair mechanisms due to internal noise, leading to cell death. However, when the Noise Controller was applied, the system successfully synchronized the cells. The failure rate dropped from 20% to 7%, dramatically increasing the survival rate. This demonstrates that mathematical control can theoretically force “lazy” or “resistant” cells to behave like the rest of the population, eliminating the outliers that typically cause treatment failure. New Era for Synthetic BiologyThis work represents a conceptual shift from population-level regulation toward single-cell precision control in stochastic biological systems. By clarifying what is mathematically achievable — and where fundamental limits lie — the study provides a foundation for future experimental and computational efforts in synthetic biology. “This research demonstrates that cellular noise — often dismissed as luck or unavoidable randomness — can be brought into the realm of precise mathematical control,” said Professor KIM Jae Kyoung, the corresponding author. “We expect this technology to play a key role in developing smart microbes and overcoming drug resistance in cancer therapy.” Professor KIM Jinsu, co-corresponding author, added, “This achievement shows the power of mathematical modeling, starting from theoretical equations to design a mechanism that solves a fundamental biological problem.” The study was published in Nature Communications.
Notes for editors
- References
- Media Contact
- About the Institute for Basic Science (IBS) |
| before |
|---|
- Content Manager
- Public Relations Team : Yim Ji Yeob 042-878-8173
- Last Update 2023-11-28 14:20