Intracellular dynamics team
Development of reduction methods for multi-scale stochastic systems
When biochemical reactions occur on disparate timescales, the Quasi-steady-state approximation (QSSA) or singular perturbation theory (e.g. Tikhonov-Fenichel theorem) utilizes timescale separation to project deterministic systems (e.g. ODE) onto lower-dimensional slow manifolds. Through this projection, biochemical reactions on slow timescales can become tractable without explicitly modeling fast reactions. This facilitates analysis and reduces computational cost. Similar to the ODE, in the presence of timescale separation, the chemical master equation (CME) can also be simplified by replacing the fast species with their QSS: the conditional average of the species given the state of the slow species [3-6]. However, so far the calculation of the conditional average is possible only in very limited cases such as linear systems, birth-death processes, feed-forward networks and complex balanced networks. We aim to extend the class of biochemical reaction networks whose stochastic QSS can be exactly derived and develop algorithms calculating the stochastic QSS with symbolic calculation. Furthermore, we aim to apply the reduction methods to accurately predict metabolism of drugs.
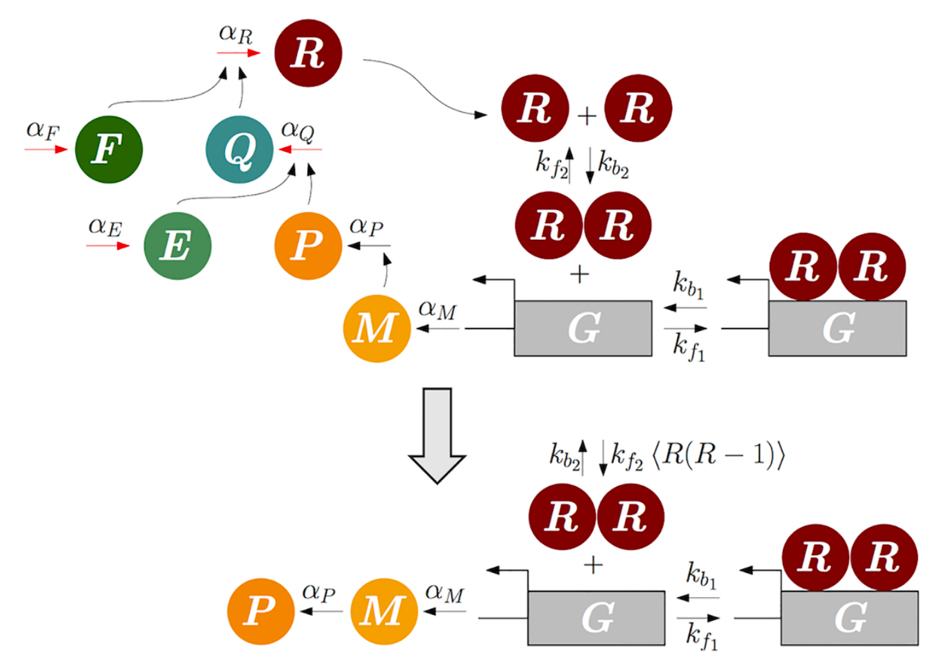
- Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: the rate-limiting step number, Science Advances (2022)
- Hierarchical Bayesian models of transcriptional and translational regulation processes with delays, Bioinformatics (2021)
- Universally valid reduction of multiscale stochastic biochemical systems using simple non-elementary propensities, PLoS Comput Biol (2021)
- Derivation of stationary distributions of biochemical reaction networks via structure transformation, Communication Biology (2021)
- Misuse of the Michaelis-Menten rate law for protein-interaction networks and its remedy, PLoS Comp Biol (2020)
- Beyond the Michaelis-Menten: Prediction of in vivo clearance for drugs with low KM, Clin Transl Sci (2020)
- Beyond the Michaelis-Menten equation: Accurate and efficient estimation of enzyme kinetic parameters, Sci Rep (2017)
- Reduction For Stochastic Biochemical Reaction Networks with Multiscale Conservations, SIAM Multiscale Model Sim (2017)
- Reduction of Multiscale Stochastic Biochemical Reaction Networks using Exact Moment Derivation, PLoS Comp Biol (2017)
- The relationship between deterministic and stochastic quasi-steady-state approximation, BMC Syst Biol (2015)
- The validity of quasi-steady-state approximations in discrete stochastic simulations, Biophy J (2014)
Identification of molecular mechanisms underlying robust circadian rhythms
The 2017 Nobel Prize in Medicine was awarded to the discovery of how cells generate sustained circadian (~24hr) rhythms via a transcriptional-translational negative feedback loop. When the repressor gene is activated by the binding of the activator, the transcription of repressor mRNA is triggered in the nucleus. Then the repressor mRNA is exported to the cytoplasm and translated to the repressor protein. Then, the repressor protein travels through the crowded cytoplasm to enter the nucleus, where the repressor inhibits its own transcriptional activator. Despite the noise in the molecular interactions and molecular movement, thousands of repressors arrive at the nucleus at the right time every day, leading to regular circadian rhythms. We aim to identify molecular mechanisms for robust circadian rhythms against spatio-temporal noise, genetic mutation, and environmental change by using the combination of mathematical modeling and experiments.
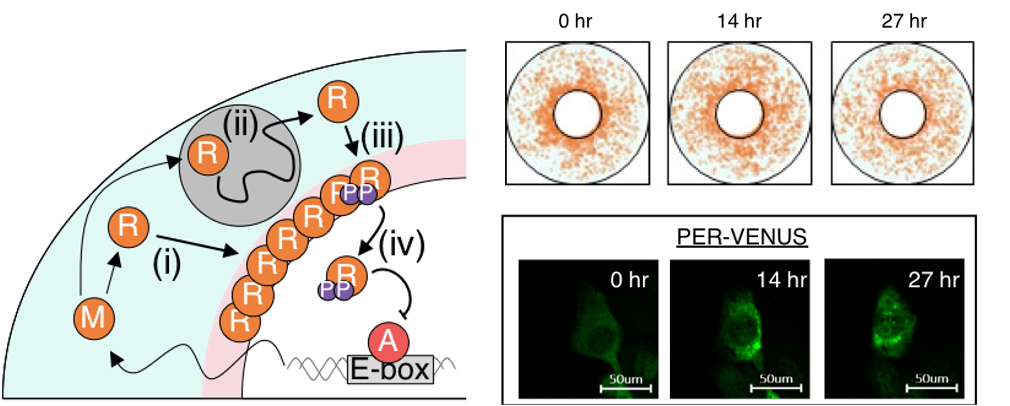
- Combined multiple transcriptional repression mechanisms generate ultrasensitivity and oscillations, Accepted for Interface Focus (2022)
- Systematic modeling-driven experiments identify distinct molecular clockworks underlying hierarchically organized pacemaker neurons, PNAS (2022)
- Modeling of Plant Circadian Clock for Characterizing Hypocotyl Growth under Different Light Quality Conditions, in silico Plants (2022)
- Wake-sleep cycles are severely disrupted by diseases affecting cytoplasmic homeostasis, PNAS (2020)
- Mutation of a PER2 phosphodegron perturbs the circadian phosphoswitch, PNAS (2020)
- Long-range temporal coordination of gene expression in spatially extended synthetic microbial consortia, Nature Chem Biol (2019)
- Distinct control of PERIOD2 degradation and circadian rhythms by the oncoprotein and ubiquitin ligase MDM2, Science Signal (2018)
- Modeling reveals a key mechanism for light-dependent phase shifts of Neurospora circadian rhythms, Biophy J (2018)
- CK1δ/ε protein kinase primes the PER2 circadian phosphoswitch. PNAS (2018)
- Stability of wake-sleep cycles requires robust degradation of the PERIOD protein, Current Biol (2017)
- Model-driven experimental approach reveals the complex regulatory distribution of p53 by the circadian factor Period 2, PNAS (2016)
- Protein sequestration versus Hill-type repression in circadian clock models, IET Syst Biol (2016)
- A tunable artificial circadian clock in clock-defective mice, Nature Commun (2015)
- Period2 Phosphoswitch Regulates and Compensates Circadian Period, Mole Cell (2015)
- Emergent genetic oscillations in a synthetic microbial consortium, Science (2015)
- Mechanism for Robust Circadian Timekeeping via stoichiometric balance, Mole Syst Biol (2012)
Intercellular dynamics team
Development of inference methods for dynamic networks
Biological systems are dynamic networks of interacting molecules and cells that regulate living specimens. Identifying the underlying dynamic networks is the first step toward understanding biological systems and disease mechanisms. However, directly measuring a network is challenging with current technology. Thus, various methods inferring the network structure from timeseries data of gene expression or protein abundance, which are easy to measure, have been developed. For instance, the causality inference method (e.g. Granger causality) and functional correlation measurement method (e.g. maximal information coefficient) have been successful in identifying network structures of various biological systems. However, these methods suffer from the false positive problem when oscillatory timeseries are given: they typically infer that two non-interacting oscillators interact with each other when they have a similar period and phase. We aim to develop network inference methods for oscillatory timeseries and for time delay distributions of the interactions.
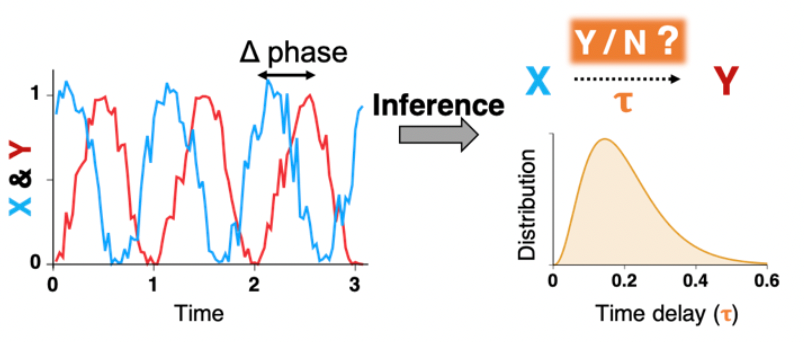
- Inference of Inferring causality in biological oscillators, Bioinformatics (2021)
- Bayesian inference of distributed time delay in transcriptional and translational regulation, Bioinformatics (2020)
- Waveforms of Molecular Oscillations Reveal Circadian Timekeeping Mechanisms, Commun Biol (2018)
- On the Existence and Uniqueness of Biological Clock Models Matching Experimental Data, SIAM J. APPL. MATH., (2012)
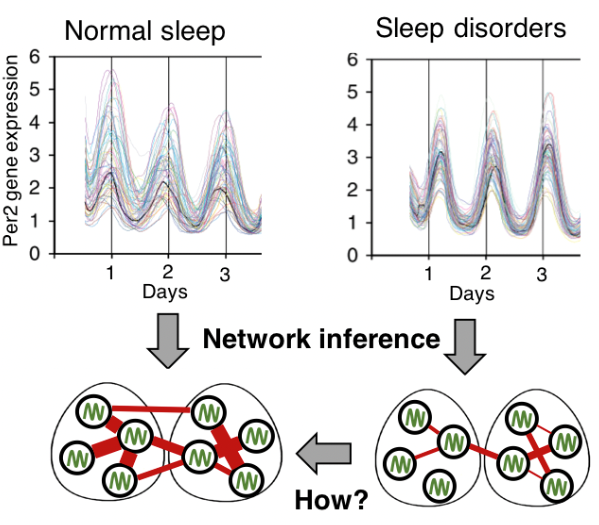
Identification of the cause of and treatment strategy for circadian rhythm sleep disorders at the network level
Circadian (~24 h) rhythms of diverse behavioral and physiological processes such as sleep and hormone secretion are coordinated by an endogenous timer, the circadian clock, which is located in the hypothalamic suprachiasmatic nucleus (SCN). The SCN consists of ~20,000 cells, each of which generates rhythmic gene expression according to its own period and phase. Intercellular coupling via various neurotransmitters (e.g. VIP and AVP) forms the dynamic network of cell populations in the SCN, which regulates phase relationship among individual rhythms and generate a global rhythm. The phase of circadian rhythms can also be adjusted by external light, which is transmitted from the retina to the SCN via the retinohypothalamic tract. This leads to the entrainment of the endogenous circadian system to the external day-night cycle. A failure of synchrony between the clock and external cycles can occur due to dysfunction of the circadian clock system or alteration of the external lighting environment, which causes circadian rhythm sleep disorders (CRSDs). Notably, more than 80% of the population appears to live a shift work lifestyle, mainly due to electrical illumination after sunset. This increases the risk for various chronic diseases such as cancer, diabetes, and mood disorders. We aim to identify the causes of and a treatment strategy for CRSDs at the network level by applying dynamic network inference algorithms to timeseries data measured from the master circadian clock.
- Molecular mechanisms that regulate the coupled period of the mammalian circadian clock, Biophy J (2014)
Systemic dynamics team
Development of methods for analyzing noisy human activity data from wearable devices and their application to identify optimal sleep patterns
About 15% of all companies and 22% of the manufacturing industry use a shift system even though the survey did not include occupational groups with relatively high potential for shift work, such as soldiers, police, health and social services workers, or part-time workers. The International Cancer Research Organization (IARC) ranked shift work at 2A, the second-highest among carcinogens. Furthermore, many industrial accidents are caused by sleepiness during shift work. We aim to develop an analysis platform for noisy and dynamic sleep patterns from wearable devices and use it to find individualized optimal sleep patterns of shift workers to minimize sleep disorders.
- Personalized sleep-wake patterns aligned with circadian rhythm relieve daytime sleepiness (submitted)
Development of chronotherapy for sleep disorders and cancer
Chronotherapy is a pharmaceutical intervention that considers the patient’s internal circadian time to adjust dosing time. Although it can dramatically improve drug efficacy and reduce toxicity, the large variability in internal time across and within individuals has prevented chronotherapies from progressing beyond clinical trials. To translate chronotherapy developments into a real-world outpatient clinical scenario, a personalized characterization and analysis of a patient’s internal time are essential. Wearable technology is expected to enable real-time high-resolution tracking of circadian and ultradian rhythms. We aim to integrate wearable data into analysis platforms including systems modeling and machine learning to pave the way toward personalized adaptive chronotherapy.
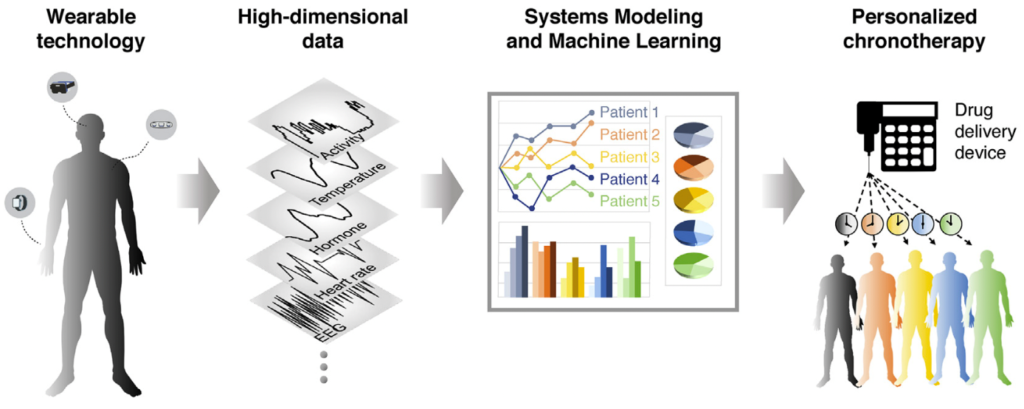
- Personalized sleep-wake patterns aligned with circadian rhythm relieve daytime sleepiness, iScience (2021)
- Combined unsupervised‑supervised machine learning for phenotyping complex diseases with its application to obstructive sleep apnea, Scientific Reports (2021)
- Wearable technology and systems modeling for personalized chronotherapy, Curr Opin Syst Biol (2020)
- Systems approach reveals photosensitivity and PER2 level as determinants of clock‐modulator efficacy, Mole Syst Biol (2019)
- Modeling and Validating Chronic Pharmacological Manipulation of Circadian Rhythms, CPT: Pharmacometrics & Systems Pharmacology (2013)

