주메뉴
- About IBS 연구원소개
-
Research Centers
연구단소개
- Research Outcomes
- Mathematics
- Physics
- Center for Theoretical Physics of the Universe(Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe(Cosmology, Gravity and Astroparticle Physics Group)
- Center for Exotic Nuclear Studies
- Center for Artificial Low Dimensional Electronic Systems
- Center for Underground Physics
- Center for Axion and Precision Physics Research
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Van der Waals Quantum Solids
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research(Neuro Technology Group)
- Center for Neuroscience Imaging Research(Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Genome Engineering
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Center for Quantum Conversion Research
- Institutes
- Korea Virus Research Institute
- News Center 뉴스 센터
- Career 인재초빙
- Living in Korea IBS School-UST
- IBS School 윤리경영


주메뉴
- About IBS
-
Research Centers
- Research Outcomes
- Mathematics
- Physics
- Center for Theoretical Physics of the Universe(Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe(Cosmology, Gravity and Astroparticle Physics Group)
- Center for Exotic Nuclear Studies
- Center for Artificial Low Dimensional Electronic Systems
- Center for Underground Physics
- Center for Axion and Precision Physics Research
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Van der Waals Quantum Solids
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Center for Neuroscience Imaging Research(Neuro Technology Group)
- Center for Neuroscience Imaging Research(Cognitive and Computational Neuroscience Group)
- Center for Algorithmic and Robotized Synthesis
- Center for Genome Engineering
- Center for Nanomedicine
- Center for Biomolecular and Cellular Structure
- Center for 2D Quantum Heterostructures
- Center for Quantum Conversion Research
- Institutes
- Korea Virus Research Institute
- News Center
- Career
- Living in Korea
- IBS School
News Center
| Title | Machine learning reveals sources of heterogeneity among cells in our bodies | ||
|---|---|---|---|
| Embargo date | 2024-01-17 15:24 | Hits | 298 |
| Press release |
![docx 파일명 : [v3] TalkFile_patterns_해외홍보_v2.docx.docx](/images/mimetype/docx.gif) [v3] TalkFile_patterns_해외홍보_v2.docx.docx
[v3] TalkFile_patterns_해외홍보_v2.docx.docx
|
||
| att. | |||
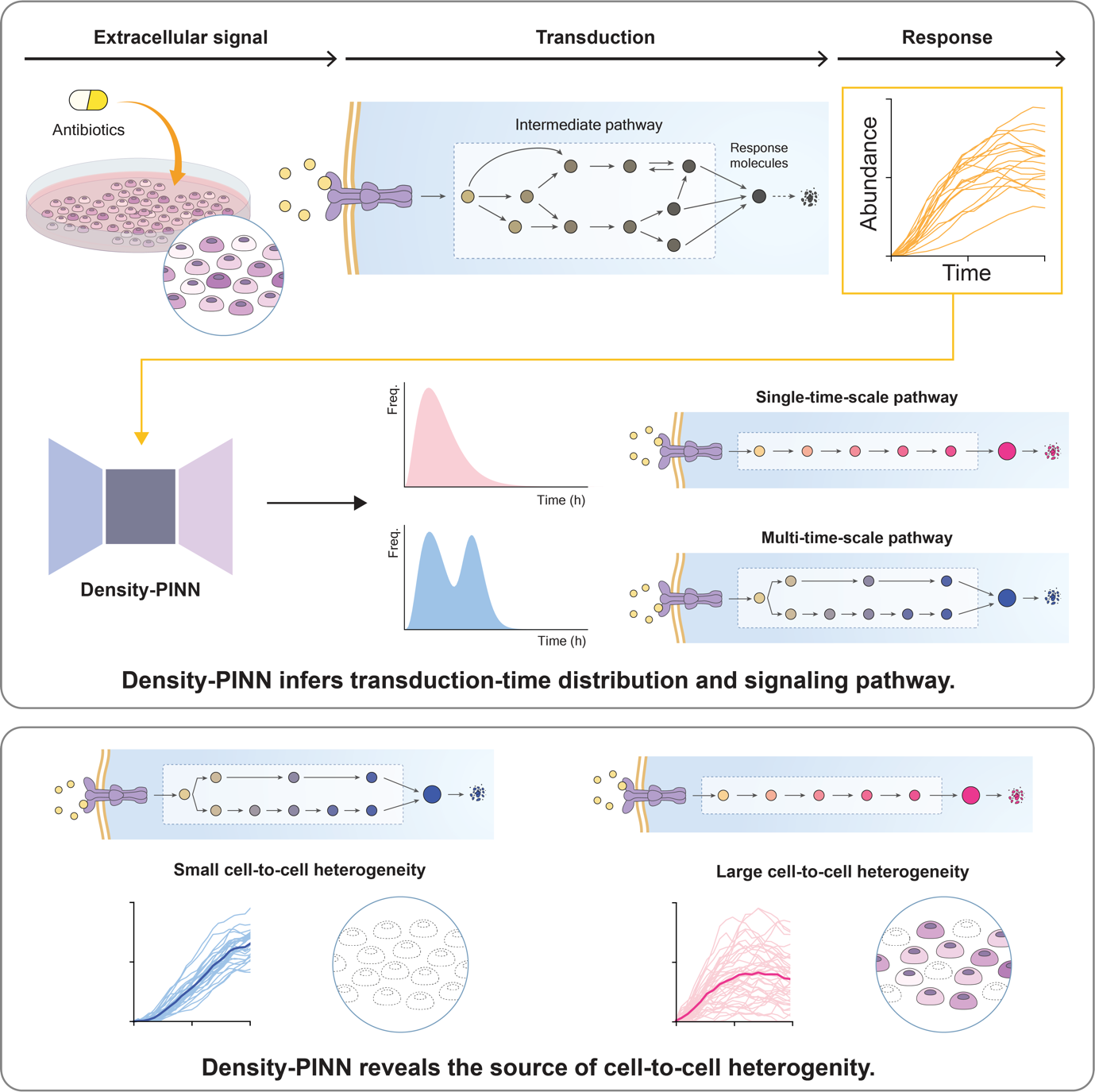
Machine learning reveals sources of heterogeneity among cells in our bodiesA team of South Korean scientists led by Professor KIM Jae Kyoung of the Biomedical Mathematics Group within the Institute for Basic Science (IBS-BIMAG) discovered the secrets of cell variability in our bodies. The findings of this research are expected to have far-reaching effects, such as improvement in the efficacy of chemotherapy treatments, or set a new paradigm in the study of antibiotic-resistant bacteria. The cells in our body have a signaling system that responds to various external stimuli such as antibiotics and osmotic pressure changes. This signaling system plays a critical role in the survival of cells as they interact with the external environment. However, even cells with same genetic information can respond differently to the same external stimuli, called cellular heterogeneity. Cellular heterogeneity is a great research interest in medicine, as it is known to hinder the complete eradication of cancer cells by chemotherapeutic agents such as anticancer drugs. The sources of such heterogeneity and its relationship with the signaling system have remained a challenge, as intermediate processes of the signaling system are impossible to fully observe with current experimental technology. To reveal the sources of this heterogeneity, Professor Kim’s research team developed a machine learning methodology using artificial neural network structures called Density Physics-informed neural networks (Density-PINNs). Density-PINNs use the observable time-series data of cells’ responses to external stimuli to inversely estimate information about the signaling system. By applying Density-PINNs to actual experimental data of antibiotic responses of bacterial cells (Escherichia coli), the research team found that a parallel structure of the signaling system can reduce heterogeneity among cells (Fig. 1). Professor Kim believes that this mathematical modeling and machine learning research will facilitate the enhancement of the understanding of cellular heterogeneity, which is crucial in cancer treatment. He expressed his hope that this achievement would lead to the development of improved cancer treatment strategies. Dr. JO Hyeontae and Dr. HONG Hyukpyo participated as co-first authors in this research, which was published in the international journal Patterns (Impact Factor 6.5), a sister journal of Cell. The title of the paper is “Density Physics-informed Neural Networks Reveal Sources of Cell Heterogeneity in Signal Transduction.”
Notes for editors
- References
- Media Contact
- About the Institute for Basic Science (IBS)
- About the Biomedical Mathematics Group (BIMAG)
|
|||
| before | |
|---|---|
| before |
- Content Manager
- Communications Team : Kwon Ye Seul 042-878-8237
- Last Update 2023-11-28 14:20