주메뉴
- About IBS 연구원소개
-
Research Centers
연구단소개
- Research Outcomes
- Mathematics
- Physics
- Center for Theoretical Physics of the Universe(Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe(Cosmology, Gravity and Astroparticle Physics Group)
- Center for Exotic Nuclear Studies
- Center for Artificial Low Dimensional Electronic Systems
- Center for Underground Physics
- Center for Axion and Precision Physics Research
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Van der Waals Quantum Solids
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Institutes
- Korea Virus Research Institute
- News Center 뉴스 센터
- Career 인재초빙
- Living in Korea IBS School-UST
- IBS School 윤리경영


주메뉴
- About IBS
-
Research Centers
- Research Outcomes
- Mathematics
- Physics
- Center for Theoretical Physics of the Universe(Particle Theory and Cosmology Group)
- Center for Theoretical Physics of the Universe(Cosmology, Gravity and Astroparticle Physics Group)
- Center for Exotic Nuclear Studies
- Center for Artificial Low Dimensional Electronic Systems
- Center for Underground Physics
- Center for Axion and Precision Physics Research
- Center for Theoretical Physics of Complex Systems
- Center for Quantum Nanoscience
- Center for Van der Waals Quantum Solids
- Chemistry
- Life Sciences
- Earth Science
- Interdisciplinary
- Institutes
- Korea Virus Research Institute
- News Center
- Career
- Living in Korea
- IBS School
News Center
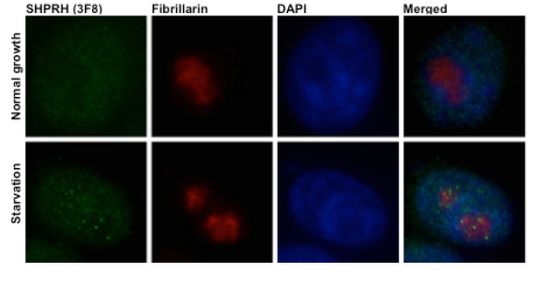
New Protein Regulated by Cellular Starvation- IBS biologists discover a new and unexpected function of a DNA repair protein - Researchers at the Center of Genomic Integrity, within the Institute for Basic Science (IBS), have found out an unexpected role for a protein involved in the DNA repair mechanism. This protein called SHPRH not only helps fixing damages during DNA replication, but also contributes to the generation of new ribosomes, the cell's "protein factories". The newly discovered task depends on the nutrition state of the cell and might be associated with aging and anemia. This research has been recently published in the Proceedings of the National Academy of Sciences (PNAS). "Some proteins that repair DNA damages are expressed at high levels by the cells even when there are no DNA mistakes to correct," explains MYUNG Kyungjae, corresponding author of the study. "Therefore it is likely that they have another role beyond DNA repair". It turns out that SHPRH is one of them. A clue about the extra function of SHPRH came from its location in the cell. IBS researchers identified where SHRPH localizes inside mammalian cells. They discovered that SHPRH is located inside the cell's nucleus, in particular in the sections of the nucleus where ribosomes are made, known as nucleoli. Interestingly, clusters of SHPRH were visible in the nucleoli after growing the cells in the same medium for some days, that is when the nutrients in the cellular medium were reduced. This triggered the idea to test SHPRH behavior during cellular starvation. As expected, the research team found that just two hours after removing nutrients from the medium, SHPRH clusters (or foci) appeared, and when nutrients were added, they disappeared again.
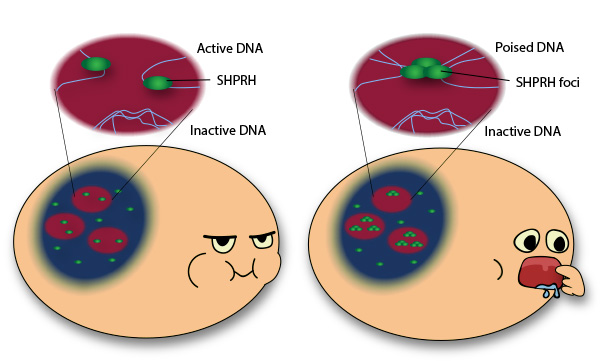
Since the nucleoli are the production sites of ribosomes, IBS scientists investigate SHPRH role in ribosome synthesis and clarified that the second function of SHPRH in mammals is associated with the transcription of rRNA, an essential component of ribosomes. "DNA repair proteins are often connected with the growth function. In this case for example, the rate of cell proliferation depends on the rate of protein synthesis, which is regulated by the ribosome and the rRNA," points out LEE Deokjae, first author of the study. Since flaws in ribosome biogenesis are connected with several metabolic problems, aging, anemia and cancers, studying these proteins is very promising. "When cells lack nutrition, they cannot grow normally, slow their energy consumption and reduce ribosome production". In case of cellular starvation, SHPRH is clustered in foci and dissociated from ribosomal DNA that is packed in a so-called 'poised' state, ready to be active as soon as the cell has enough nutrients. For this reason, if nutrients are added during the starvation experiment, the SHPRH spots disappear and the ribosome production can be quickly recovered.
"We have yet to clarify the exact role of these SHPRH foci," comments Lee. "Are they just a temporary trash-bin of unnecessary proteins? Are they connected with aging? We still do not know, but we have several hypothesis. Future experiments will tell". Letizia Diamante Notes for editors - References - Media Contact - About the Institute for Basic Science (IBS) |
|||
Center for Genomic Integrity |
|||
|
|
| Next | |
|---|---|
| before |
- Content Manager
- Public Relations Team : Suh, William Insang 042-878-8137
- Last Update 2023-11-28 14:20